7QKB
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound GC376 | | Descriptor: | CHLORIDE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
2PWG
 
 | | Crystal Structure of the Trehalulose Synthase MutB From Pseudomonas Mesoacidophila MX-45 Complexed to the Inhibitor Castanospermine | | Descriptor: | CALCIUM ION, CASTANOSPERMINE, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
9C5S
 
 | | Disulfide-linked, antiparallel p53-derived peptide dimer (CV1) | | Descriptor: | Cellular tumor antigen p53, SULFATE ION | | Authors: | Vithanage, N, Kreitler, D.K, DiGiorno, M.C, Victorio, C.G, Sawyer, N, Outlaw, V.K. | | Deposit date: | 2024-06-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structural Characterization of Disulfide-Linked p53-Derived Peptide Dimers.
Res Sq, 2024
|
|
7QKA
 
 | | Crystal structure of SARS-CoV-2 Main Protease in complex with covalently bound GC376 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
1QG0
 
 | | WILD-TYPE PEA FNR | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE) | | Authors: | Deng, Z, Aliverti, A, Zanetti, G, Arakaki, A.K, Ottado, J, Orellano, E.G, Calcaterra, N.B, Ceccarelli, E.A, Carrillo, N, Karplus, P.A. | | Deposit date: | 1999-04-18 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A productive NADP+ binding mode of ferredoxin-NADP+ reductase revealed by protein engineering and crystallographic studies.
Nat.Struct.Biol., 6, 1999
|
|
1QGA
 
 | | PEA FNR Y308W MUTANT IN COMPLEX WITH NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), ... | | Authors: | Deng, Z, Aliverti, A, Zanetti, G, Arakaki, A.K, Ottado, J, Orellano, E.G, Calcaterra, N.B, Ceccarelli, E.A, Carrillo, N, Karplus, P.A. | | Deposit date: | 1999-04-18 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A productive NADP+ binding mode of ferredoxin-NADP+ reductase revealed by protein engineering and crystallographic studies.
Nat.Struct.Biol., 6, 1999
|
|
1QKF
 
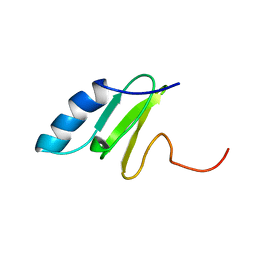 | | SOLUTION STRUCTURE OF THE RIBOSOMAL PROTEIN S19 FROM THERMUS THERMOPHILUS | | Descriptor: | 30S RIBOSOMAL PROTEIN S19 | | Authors: | Helgstrand, M, Rak, A.V, Allard, P, Davydova, N, Garber, M.B, Hard, T. | | Deposit date: | 1999-07-19 | | Release date: | 1999-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosomal protein S19 from Thermus thermophilus.
J. Mol. Biol., 292, 1999
|
|
1QOJ
 
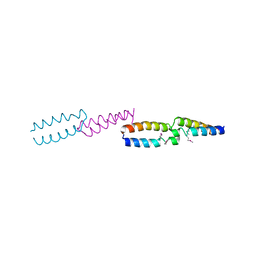 | | Crystal Structure of E.coli UvrB C-terminal domain, and a model for UvrB-UvrC interaction. | | Descriptor: | UVRB | | Authors: | Sohi, M, Alexandrovich, A, Moolenaar, G, Visse, R, Goosen, N, Vernede, X, Fontecilla-Camps, J, Champness, J, Sanderson, M.R. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-10 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of E.Coli Uvrb C-Terminal Domain, and a Model for Uvrb-Uvrc Interaction
FEBS Lett., 465, 2000
|
|
2PUM
 
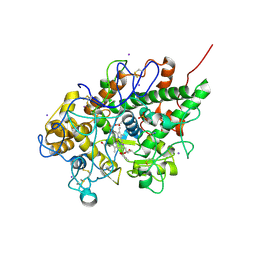 | | Crystal structure of bovine lactoperoxidase complex with catechol and iodide at 2.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CATECHOL, ... | | Authors: | Singh, A.K, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-05-09 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of bovine lactoperoxidase complex with catechol and iodide at 2.7 A resolution
To be Published
|
|
2PUS
 
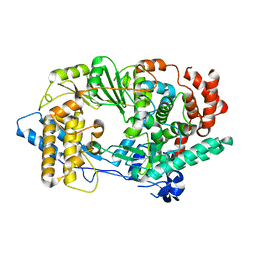 | | Unprecedented activation mechanism of a non-canonical RNA-dependent RNA polymerase | | Descriptor: | IBDV VP1 RNA-dependant RNA polymerase | | Authors: | Garriga, D, Navarro, A, Querol-Audi, J, Abaitua, F, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2007-05-09 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation mechanism of a noncanonical RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1QVV
 
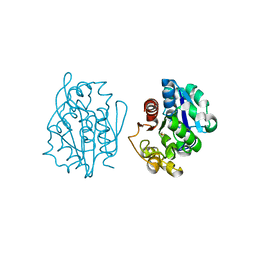 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1QOX
 
 | |
2PWE
 
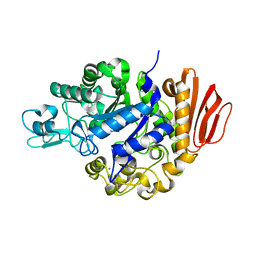 | | Crystal structure of the MutB E254Q mutant in complex with the substrate sucrose | | Descriptor: | CALCIUM ION, Sucrose isomerase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 282, 2007
|
|
2PWA
 
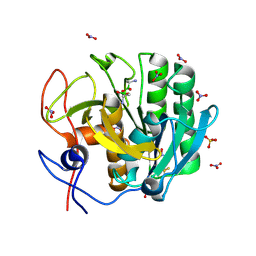 | | Crystal Structure of the complex of Proteinase K with Alanine Boronic acid at 0.83A resolution | | Descriptor: | ALANINE BORONIC ACID, CALCIUM ION, NITRATE ION, ... | | Authors: | Jain, R, Singh, N, Perbandt, M, Betzel, C, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-05-11 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Crystal structure of the complex of Proteinase K with Alanine Boronic Acid at 0.83A Resolution
To be Published
|
|
2PY4
 
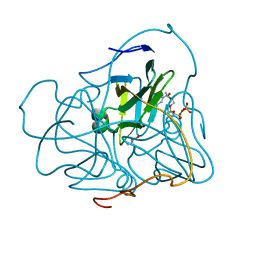 | | Full length structure of the Mycobacterium tuberculosis dUTPase complexed with magnesium and alpha,beta-imido-dUTP. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Nagy, N, Takacs, E, Vertessy, B.G. | | Deposit date: | 2007-05-15 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Active site of mycobacterial dUTPase: structural characteristics and a built-in sensor.
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2PYC
 
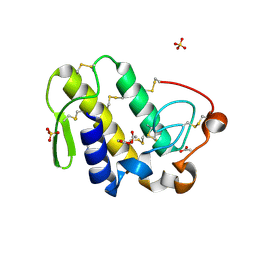 | | Crystal structure of a monomeric phospholipase A2 from Russell's viper at 1.5A resolution | | Descriptor: | ACETATE ION, ACETONITRILE, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2007-05-16 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a monomeric phospholipase A2 from Russell's viper at 1.5A resolution
To be Published
|
|
1Q6V
 
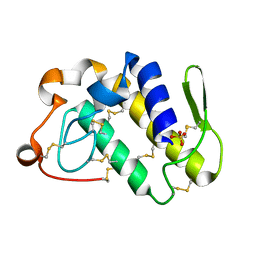 | | First crystal structure of a C49 monomer PLA2 from the venom of Daboia russelli pulchella at 1.8 A resolution | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Pal, A, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-08-14 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | First crystal structure of a C49 PLA2 from the venom of Daboia russelli pulchella at 1.8A resolution
To be Published
|
|
1QDD
 
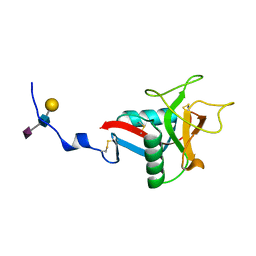 | | CRYSTAL STRUCTURE OF HUMAN LITHOSTATHINE TO 1.3 A RESOLUTION | | Descriptor: | LITHOSTATHINE, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Gerbaud, V, Pignol, D, Loret, E, Bertrand, J.A, Berland, Y, Fontecilla-Camps, J.C, Canselier, J.P, Gabas, N, Verdier, J.M. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of calcite crystal growth inhibition by the N-terminal undecapeptide of lithostathine.
J.Biol.Chem., 275, 2000
|
|
2Q6A
 
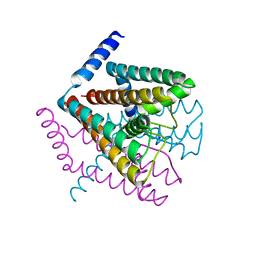 | | Crystal Structure of Nak channel D66E mutant | | Descriptor: | CALCIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Alam, A, Shi, N, Jiang, Y. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into Ca2+ specificity in tetrameric cation channels.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1QVP
 
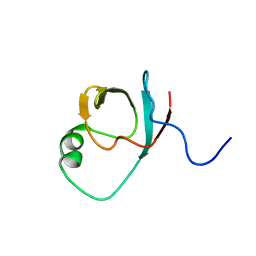 | | C terminal SH3-like domain from Diphtheria toxin Repressor residues 144-226. | | Descriptor: | Diphtheria toxin repressor | | Authors: | Wylie, G.P, Rangachari, V, Bienkiewicz, E.A, Marin, V, Bhattacharya, N, Love, J.F, Murphy, J.R, Logan, T.M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch.
Biochemistry, 44, 2005
|
|
2PWH
 
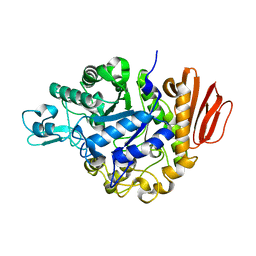 | | Crystal structure of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45 | | Descriptor: | CALCIUM ION, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
8CRF
 
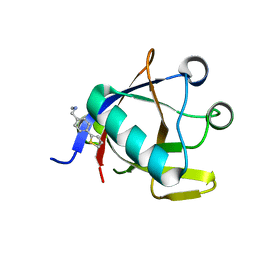 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 5E11 refined against anomalous diffraction data | | Descriptor: | Host translation inhibitor nsp1, ~{N}-methyl-1-(4-thiophen-2-ylphenyl)methanamine | | Authors: | Ma, S, Mykhaylyk, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8CRK
 
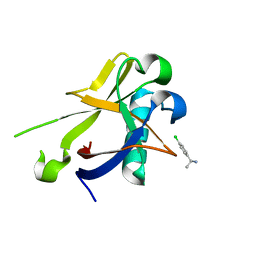 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2 refined against anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-chlorophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8CRM
 
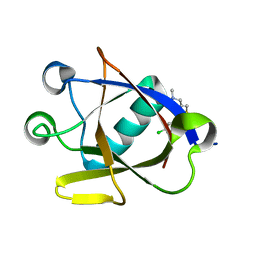 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11C6 refined against anomalous diffraction data | | Descriptor: | 1-[2-(3-chlorophenyl)-1,3-thiazol-4-yl]-~{N}-methyl-methanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
2Q2W
 
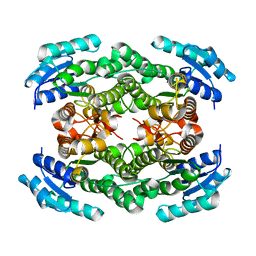 | | Structure of D-3-Hydroxybutyrate Dehydrogenase from Pseudomonas putida | | Descriptor: | Beta-D-hydroxybutyrate dehydrogenase | | Authors: | Paithankar, K.S, Feller, C, Kuettner, E.B, Keim, A, Grunow, M, Strater, N. | | Deposit date: | 2007-05-29 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Cosubstrate-induced dynamics of D-3-hydroxybutyrate dehydrogenase from Pseudomonas putida.
Febs J., 274, 2007
|
|
