2QJO
 
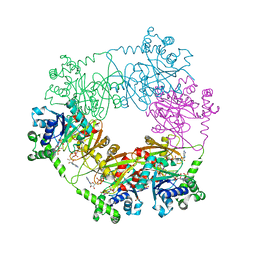 | | crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase (NadM) complexed with ADPRP and NAD from Synechocystis sp. | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Bifunctional NMN adenylyltransferase/Nudix hydrolase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-08 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
1M5T
 
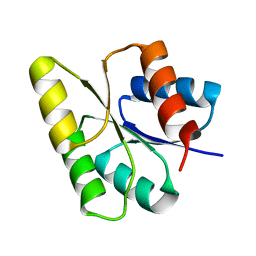 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK | | Descriptor: | cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic and biochemical studies of DivK reveal novel features of an essential response regulator in Caulobacter crescentus
J.Biol.Chem., 277, 2002
|
|
1MB0
 
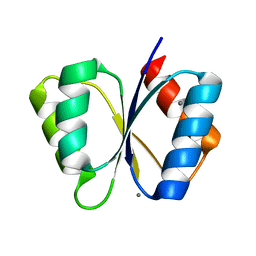 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 8.0 IN COMPLEX WITH MN2+ | | Descriptor: | MANGANESE (II) ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
5AQI
 
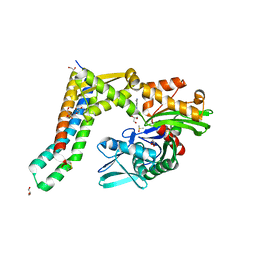 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | ADENINE, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, DIMETHYL SULFOXIDE, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AQK
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, DIMETHYL SULFOXIDE, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AQQ
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7-methylquinazolin-4-amine, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
1N96
 
 | | DIMERIC SOLUTION STRUCTURE OF THE CYCLIC OCTAMER CD(CGCTCATT) | | Descriptor: | CYCLIC OLIGONUCLEOTIDE D(CGCTCATT) | | Authors: | Escaja, N, Gelpi, J.L, Orozco, M, Rico, M, Pedroso, E, Gonzalez, C. | | Deposit date: | 2002-11-22 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Four-stranded DNA structure stabilized by a novel G:C:A:T tetrad.
J.Am.Chem.Soc., 125, 2003
|
|
1KHO
 
 | | Crystal Structure Analysis of Clostridium perfringens alpha-Toxin Isolated from Avian Strain SWCP | | Descriptor: | ZINC ION, alpha-toxin | | Authors: | Justin, N, Moss, D.S, Titball, R.W, Basak, A.K. | | Deposit date: | 2001-11-30 | | Release date: | 2002-06-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The first strain of Clostridium perfringens isolated from an avian source has an alpha-toxin with divergent structural and kinetic properties.
Biochemistry, 41, 2002
|
|
1OSI
 
 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
1MXF
 
 | | Crystal Structure of Inhibitor Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
8JQO
 
 | | Protocatecuate hydroxylase from Xylophilus ampelinus complexed with imidazole | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
5LGQ
 
 | | Crystal structure of mouse CARM1 in complex with ligand P2C3s | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-propyl-oxolane-3,4-diol, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Marechal, N, Troffer-Charlier, N, Cura, V, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-07-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Transition state mimics are valuable mechanistic probes for structural studies with the arginine methyltransferase CARM1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1WYT
 
 | | Crystal structure of glycine decarboxylase (P-protein) of the glycine cleavage system, in apo form | | Descriptor: | glycine dehydrogenase (decarboxylating) subunit 1, glycine dehydrogenase subunit 2 (P-protein) | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of P-protein of the glycine cleavage system: implications for nonketotic hyperglycinemia
Embo J., 24, 2005
|
|
6P5S
 
 | | HIPK2 kinase domain bound to CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Homeodomain-interacting protein kinase 2 | | Authors: | Agnew, C, Liu, L, Jura, N. | | Deposit date: | 2019-05-30 | | Release date: | 2019-07-31 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | The crystal structure of the protein kinase HIPK2 reveals a unique architecture of its CMGC-insert region.
J.Biol.Chem., 294, 2019
|
|
5LGS
 
 | | Crystal structure of mouse CARM1 in complex with ligand P2C3u | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Marechal, N, Troffer-Charlier, N, Cura, V, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-07-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transition state mimics are valuable mechanistic probes for structural studies with the arginine methyltransferase CARM1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5FUB
 
 | | Crystal Structure of zebrafish Protein Arginine Methyltransferase 2 catalytic domain with SAH | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-01-22 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
5FUL
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with SAH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
5MI8
 
 | | Structure of the phosphomimetic mutant of EF-Tu T383E | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Talavera, A, Hendrix, J, Versees, W, De Gieter, S, Castro-Roa, D, Jurenas, D, Van Nerom, K, Vandenberk, N, Barth, A, De Greve, H, Hofkens, J, Zenkin, N, Loris, R, Garcia-Pino, A. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors.
Sci Adv, 4, 2018
|
|
8KG3
 
 | | Structure of THOUSAND-GRAIN WEIGHT 6 (TGW6) | | Descriptor: | Os06g0623700 protein | | Authors: | Akabane, T, Suzuki, N, Matsumura, H, Yoshizawa, T, Tsuchiya, W, Katoh, E, Hirotsu, N. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | THOUSAND-GRAIN WEIGHT 6, which is an IAA-glucose hydrolase, preferentially recognizes the structure of the indole ring.
Sci Rep, 14, 2024
|
|
5FWD
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP2 | | Descriptor: | 1,2-ETHANEDIOL, 9-(7-{[amino(iminio)methyl]amino}-5,6,7-trideoxy-beta-D-ribo-heptofuranosyl)-9H-purin-6-amine, CALCIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-02-12 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies of Protein Arginine Methyltransferase 2 Reveal its Interactions with Potential Substrates and Inhibitors.
FEBS J., 284, 2017
|
|
5GTF
 
 | | Crystal structure of onion lachrymatory factor synthase (LFS) containing glycerol | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lachrymatory-factor synthase, ... | | Authors: | Takabe, J, Arakawa, T, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2016-08-20 | | Release date: | 2017-08-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase
Acs Catalysis, 10, 2020
|
|
7Y8P
 
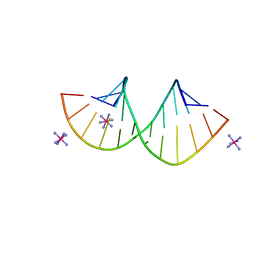 | | Crystal structure of 4'-selenoRNA duplex | | Descriptor: | COBALT HEXAMMINE(III), RNA (5'-R(*GP*GP*AP*(IKS)P*(ILK)P*(IKS)P*GP*AP*GP*UP*CP*C)-3') | | Authors: | Kondo, J, Minakawa, N, Ohta, M, Takahashi, H, Tarashima, N. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and properties of fully-modified 4'-selenoRNA, an endonuclease-resistant RNA analog.
Bioorg.Med.Chem., 76, 2022
|
|
8H8C
 
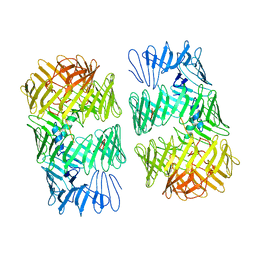 | | Type VI secretion system effector RhsP in its post-autoproteolysis and dimeric form | | Descriptor: | C-terminal peptide from Putative Rhs-family protein, Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
8H8B
 
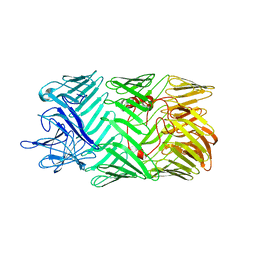 | | Type VI secretion system effector RhsP in its pre-autoproteolysis and monomeric form | | Descriptor: | Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
5GTE
 
 | | Crystal structure of onion lachrymatory factor synthase (LFS), solute-free form | | Descriptor: | Lachrymatory-factor synthase, SULFATE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Takabe, J, Arakawa, T, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2016-08-20 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase
Acs Catalysis, 10, 2020
|
|
