2WVK
 
 | | Mannosyl-3-phosphoglycerate synthase from Thermus thermophilus HB27 apoprotein | | Descriptor: | CITRATE ANION, MANNOSYL-3-PHOSPHOGLYCERATE SYNTHASE, ZINC ION | | Authors: | Goncalves, S, Borges, N, Esteves, A.M, Victor, B, Soares, C.M, Santos, H, Matias, P.M. | | Deposit date: | 2009-10-19 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural Analysis of Thermus Thermophilus Hb27 Mannosyl-3-Phosphoglycerate Synthase Provides Evidence for a Second Catalytic Metal Ion and New Insight Into the Retaining Mechanism of Glycosyltransferases.
J.Biol.Chem., 285, 2010
|
|
3ANS
 
 | | Human soluble epoxide hydrolase in complex with a synthetic inhibitor | | Descriptor: | 4-cyano-N-[(1S,2R)-2-phenylcyclopropyl]benzamide, Epoxide hydrolase 2 | | Authors: | Chiyo, N, Ishii, T, Hourai, S, Yanagi, K. | | Deposit date: | 2010-09-08 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A practical use of ligand efficiency indices out of the fragment-based approach: ligand efficiency-guided lead identification of soluble epoxide hydrolase inhibitors
J.Med.Chem., 54, 2011
|
|
2HLD
 
 | | Crystal structure of yeast mitochondrial F1-ATPase | | Descriptor: | ATP synthase alpha chain, mitochondrial, ATP synthase beta chain, ... | | Authors: | Kabaleeswaran, V, Puri, N, Walker, J.E, Leslie, A.G, Mueller, D.M. | | Deposit date: | 2006-07-06 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel features of the rotary catalytic mechanism revealed in the structure of yeast F(1) ATPase.
Embo J., 25, 2006
|
|
1OXF
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
7CCO
 
 | | The binding structure of a lanthanide binding tag (LBT3) with lanthanum ion (La3+) | | Descriptor: | LANTHANUM (III) ION, LBT3 | | Authors: | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | Deposit date: | 2020-06-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
1P29
 
 | | Crystal Structure of glycogen phosphorylase b in complex with maltopentaose | | Descriptor: | Glycogen phosphorylase, muscle form, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Pinotsis, N, Leonidas, D.D, Chrysina, E.D, Oikonomakos, N.G, Mavridis, I.M. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The binding of beta- and gamma-cyclodextrins to glycogen phosphorylase b: Kinetic and crystallographic studies.
Protein Sci., 12, 2003
|
|
1P7N
 
 | | Dimeric Rous Sarcoma virus Capsid protein structure with an upstream 25-amino acid residue extension of C-terminal of Gag p10 protein | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27 | | Authors: | Nandhagopal, N, Simpson, A.A, Johnson, M.C, Francisco, A.B, Schatz, G.W, Rossmann, M.G, Vogt, V.M. | | Deposit date: | 2003-05-02 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dimeric rous sarcoma virus capsid protein structure relevant to immature gag assembly
J.Mol.Biol., 335, 2004
|
|
1ONL
 
 | | Crystal structure of Thermus thermophilus HB8 H-protein of the glycine cleavage system | | Descriptor: | glycine cleavage system H protein | | Authors: | Nakai, T, Ishijima, J, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-28 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Thermus thermophilus HB8 H-protein of the glycine-cleavage system, resolved by a six-dimensional molecular-replacement method.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5B8B
 
 | | Crystal structure of reduced chimeric E.coli AhpC1-186-YFSKHN | | Descriptor: | Alkyl hydroperoxide reductase subunit C,Peroxiredoxin-2, SULFATE ION | | Authors: | Kamariah, N, Sek, M.F, Eisenhaber, B, Eisenhaber, F, Gruber, G. | | Deposit date: | 2016-06-14 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Transition steps in peroxide reduction and a molecular switch for peroxide robustness of prokaryotic peroxiredoxins.
Sci Rep, 6, 2016
|
|
6IC1
 
 | | urate oxidase under 90 bar of krypton | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-AZAXANTHINE, ACETATE ION, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
1P1N
 
 | |
1P2G
 
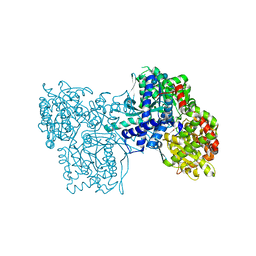 | | Crystal Structure of Glycogen Phosphorylase B in complex with Gamma Cyclodextrin | | Descriptor: | Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Glycogen phosphorylase, muscle form, ... | | Authors: | Pinotsis, N, Leonidas, D.D, Chrysina, E.D, Oikonomakos, N.G, Mavridis, I.M. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The binding of beta- and gamma-cyclodextrins to glycogen phosphorylase b: Kinetic and crystallographic studies.
Protein Sci., 12, 2003
|
|
1MHY
 
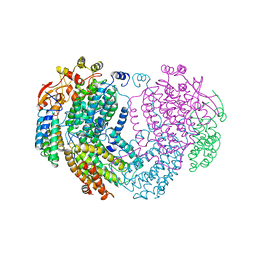 | | METHANE MONOOXYGENASE HYDROXYLASE | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Elango, N, Radhakrishnan, R, Froland, W.A, Waller, B.J, Earhart, C.A, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1996-10-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b
Protein Sci., 6, 1997
|
|
1MA8
 
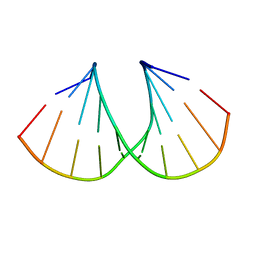 | | A-DNA decamer GCGTA(UMS)ACGC with incorporated 2'-methylseleno-uridine | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*UMSP*AP*CP*GP*C)-3' | | Authors: | Teplova, M, Wilds, C.J, Wawrzak, Z, Tereshko, V, Du, Q, Carrasco, N, Huang, Z, Egli, M. | | Deposit date: | 2002-08-01 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Covalent incorporation of selenium into oligonucleotides for X-ray crystal structure determination via MAD: proof of principle
BIOCHIMIE, 84, 2002
|
|
1MPV
 
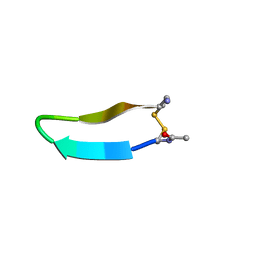 | | Structure of bhpBR3, the BAFF-binding loop of BR3 embedded in a beta-hairpin peptide | | Descriptor: | BLyS Receptor 3 | | Authors: | Kayagaki, N, Yan, M, Seshasayee, D, Wang, H, Lee, W, French, D.M, Grewal, I.S, Cochran, A.G, Gordon, N.C, Yin, J, Starovasnik, M.A, Dixit, V.M. | | Deposit date: | 2002-09-12 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kappaB2.
Immunity, 17, 2002
|
|
1M4W
 
 | | Thermophilic b-1,4-xylanase from Nonomuraea flexuosa | | Descriptor: | ACETATE ION, GLYCEROL, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hakulinen, N, Turunen, O, Janis, J, Leisola, M, Rouvinen, J. | | Deposit date: | 2002-07-05 | | Release date: | 2003-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structures of thermophilic beta-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability.
Eur.J.Biochem., 270, 2003
|
|
1M53
 
 | |
1MH2
 
 | | Crystal Structure of a Zinc Containing Dimer of Phospholipase A2 from the Venom of Indian Cobra (Naja Naja Sagittifera) | | Descriptor: | ACETIC ACID, PHOSPHOLIPASE A2, ZINC ION | | Authors: | Jabeen, T, Varma, A.K, Paramasivam, M, Singh, N, Singh, R.K, Sharma, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-08-19 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Zinc Containing Dimer of Phospholipase A2 from the Venom of Indian cobra (Naja Naja Saggittifera)
To be Published
|
|
1MDB
 
 | | CRYSTAL STRUCTURE OF DhbE IN COMPLEX WITH DHB-ADENYLATE | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase, ADENOSINE MONOPHOSPHATE, ... | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1NAV
 
 | | Thyroid Receptor Alpha in complex with an agonist selective for Thyroid Receptor Beta1 | | Descriptor: | SULFATE ION, hormone receptor alpha 1, THRA1, ... | | Authors: | Ye, L, Li, Y.L, Mellstrom, K, Mellin, C, Bladh, L.G, Koehler, K, Garg, N, Garcia Collazo, A.M, Litten, C, Husman, B, Persson, K, Ljunggren, J, Grover, G, Sleph, P.G, George, R, Malm, J. | | Deposit date: | 2002-11-29 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thyroid receptor ligands. 1. Agonist ligands selective for the thyroid receptor beta1.
J.Med.Chem., 46, 2003
|
|
1NI1
 
 | | Imidazole and cyanophenyl farnesyl transferase inhibitors | | Descriptor: | 2-CHLORO-5-(3-CHLORO-PHENYL)-6-[(4-CYANO-PHENYL)-(3-METHYL-3H-IMIDAZOL-4-YL)- METHOXYMETHYL]-NICOTINONITRILE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Tong, Y, Lin, N.H, Wang, L, Hasvold, L, Wang, W, Leonard, N, Li, T, Li, Q, Cohen, J, Gu, W.Z, Zhang, H, Stoll, V, Bauch, J, Marsh, K, Rosenberg, S.H, Sham, H.L. | | Deposit date: | 2002-12-20 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of potent imidazole and cyanophenyl containing farnesyltransferase inhibitors with improved oral bioavailability.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1N9S
 
 | | Crystal structure of yeast SmF in spacegroup P43212 | | Descriptor: | Small nuclear ribonucleoprotein F | | Authors: | Collins, B.M, Cubeddu, L, Naidoo, N, Harrop, S.J, Kornfeld, G.D, Dawes, I.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2002-11-26 | | Release date: | 2002-12-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Homomeric ring assemblies of eukaryotic Sm proteins have affinity for
both RNA and DNA: Crystal structure of an oligomeric complex of yeast
SmF
J.Biol.Chem., 278, 2003
|
|
1NI2
 
 | | Structure of the active FERM domain of Ezrin | | Descriptor: | Ezrin | | Authors: | Smith, W.J, Nassar, N, Bretscher, A.P, Cerione, R.A, Karplus, P.A. | | Deposit date: | 2002-12-20 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Active N-terminal Domain of Ezrin. CONFORMATIONAL AND MOBILITY CHANGES IDENTIFY KEYSTONE INTERACTIONS.
J.Biol.Chem., 278, 2003
|
|
1NKR
 
 | | INHIBITORY RECEPTOR (P58-CL42) FOR HUMAN NATURAL KILLER CELLS | | Descriptor: | P58-CL42 KIR | | Authors: | Fan, Q.R, Mosyak, L, Winter, C.C, Wagtmann, N, Long, E.O, Wiley, D.C. | | Deposit date: | 1998-06-24 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors.
Nature, 389, 1997
|
|
1POU
 
 | | THE SOLUTION STRUCTURE OF THE OCT-1 POU-SPECIFIC DOMAIN REVEALS A STRIKING SIMILARITY TO THE BACTERIOPHAGE LAMBDA REPRESSOR DNA-BINDING DOMAIN | | Descriptor: | OCT-1 | | Authors: | Assa-Munt, N, Mortishire-Smith, R.J, Aurora, R, Herr, W, Wright, P.E. | | Deposit date: | 1993-06-14 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage lambda repressor DNA-binding domain.
Cell(Cambridge,Mass.), 73, 1993
|
|
