4U1E
 
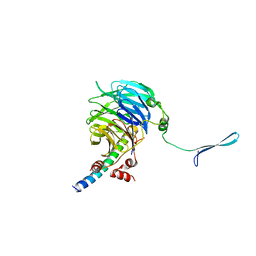 | | Crystal structure of the eIF3b-CTD/eIF3i/eIF3g-NTD translation initiation complex | | Descriptor: | Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit G, Eukaryotic translation initiation factor 3 subunit I | | Authors: | Zhang, S, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-07-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell, 158, 2014
|
|
4U52
 
 | | Crystal structure of Nagilactone C bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
3M1H
 
 | |
1P4G
 
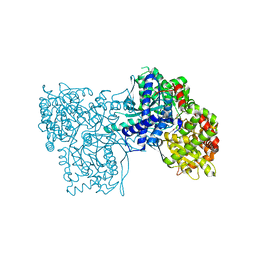 | | Crystal structure of glycogen phosphorylase b in complex with C-(1-azido-alpha-D-glucopyranosyl)formamide | | Descriptor: | C-(1-AZIDO-ALPHA-D-GLUCOPYRANOSYL) FORMAMIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Oikonomakos, N.G, Zographos, S.E, Kosmopoulou, M.N, Bischler, N, Leonidas, D.D, Nagy, V, Somsak, L, Gergely, P, Praly, J.-P. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crsytallographic Studies on alpha- and beta-D-glucopyranosyl Formamide Analogues, Inhibitors of Glycogen Phosphorylase
Biocatal.Biotransfor., 21, 2003
|
|
1OYO
 
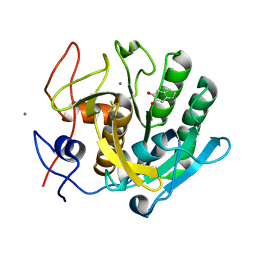 | | Regulation of protease activity by melanin: Crystal structure of the complex formed between proteinase K and melanin monomers at 2.0 resolution | | Descriptor: | 3H-INDOLE-5,6-DIOL, CALCIUM ION, Proteinase K | | Authors: | Singh, N, Sharma, S, Kumar, S, Raman, G, Singh, T.P. | | Deposit date: | 2003-04-06 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Regulation of protease activity by melanin: Crystal structure of the complex formed between proteinase K and melanin monomers at 2.0 resolution
To be Published
|
|
4UB7
 
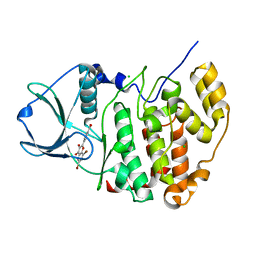 | | High-salt structure of protein kinase CK2 catalytic subunit with 4'-carboxy-6,8-bromo-flavonol (FLC26) showing an extreme distortion of the ATP-binding loop combined with a pi-halogen bond | | Descriptor: | 4-(6,8-dibromo-3-hydroxy-4-oxo-4H-chromen-2-yl)benzoic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Bischoff, N, Guerra, B, Golub, A, Issinger, O.-G. | | Deposit date: | 2014-08-12 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Note of Caution on the Role of Halogen Bonds for Protein Kinase/Inhibitor Recognition Suggested by High- And Low-Salt CK2 alpha Complex Structures.
Acs Chem.Biol., 10, 2015
|
|
5MB6
 
 | |
1OKO
 
 | | Crystal structure of Pseudomonas Aeruginosa Lectin 1 complexed with galactose at 1.6 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PA-I GALACTOPHILIC LECTIN, ... | | Authors: | Cioci, G, Mitchell, E, Gautier, C, Wimmerova, M, Perez, S, Gilboa-Garber, N, Imberty, A. | | Deposit date: | 2003-07-28 | | Release date: | 2003-12-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of Calcium and Galactose Recognition by the Lectin Pa-Il of Pseudomonas Aeruginosa
FEBS Lett., 555, 2003
|
|
4U3M
 
 | | Crystal structure of Anisomycin bound to the yeast 80S ribosome | | Descriptor: | 18S rRNA, 25s rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
1OLN
 
 | | Model for thiostrepton antibiotic binding to L11 substrate from 50S ribosomal RNA | | Descriptor: | 50S RIBOSOMAL PROTEIN L11, RNA, THIOSTREPTON | | Authors: | Lentzen, G, Klinck, R, Matassova, N, Aboul-Ela, F, Murchie, A.I.H. | | Deposit date: | 2003-08-08 | | Release date: | 2003-09-11 | | Last modified: | 2019-08-21 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Basis for Contrasting Activities of Ribosome Binding Thiazole Antibiotics
Chem.Biol., 10, 2003
|
|
1OO2
 
 | | Crystal structure of transthyretin from Sparus aurata | | Descriptor: | CADMIUM ION, transthyretin | | Authors: | Pasquato, N, Ramazzina, I, Folli, C, Battistutta, R, Berni, R, Zanotti, G. | | Deposit date: | 2003-03-03 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Distinctive binding and structural properties of piscine transthyretin.
Febs Lett., 555, 2003
|
|
6GMS
 
 | | Solution NMR structure of the major type IV pilin PpdD from enterohemorrhagic Escherichia coli (EHEC) | | Descriptor: | Prepilin peptidase-dependent protein D | | Authors: | Amorim, G.C, Bardiaux, B, Luna-Rico, A, Zeng, W, Guilvout, I, Egelman, E, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2018-05-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus.
Structure, 27, 2019
|
|
1O9S
 
 | | Crystal structure of a ternary complex of the human histone methyltransferase SET7/9 | | Descriptor: | GENE FRAGMENT FOR HISTONE H3, HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC, ... | | Authors: | Xiao, B, Jing, C, Wilson, J.R, Walker, P.A, Vasisht, N, Kelly, G, Howell, S, Taylor, I.A, Blackburn, G.M, Gamblin, S.J. | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Catalytic Mechanism of the Human Histone Methyltransferase Set7/9
Nature, 421, 2003
|
|
7F1G
 
 | | BACE2 xaperone complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-2H,3H-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Descriptor: | Beta-secretase 2, N-[3-[(4R,5R,6R)-2-azanyl-5-fluoranyl-4,6-dimethyl-5,6-dihydro-1,3-thiazin-4-yl]-4-fluoranyl-phenyl]-2,3-dihydro-[1,4]dioxino[2,3-c]pyridine-7-carboxamide, XAPERONE | | Authors: | Ueno, T, Matsuoka, E, Asada, N, Yamamoto, S, Kanegawa, N, Ito, M, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2021-06-09 | | Release date: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Extremely Selective Fused Pyridine-Derived beta-Site Amyloid Precursor Protein-Cleaving Enzyme (BACE1) Inhibitors with High In Vivo Efficacy through 10s Loop Interactions.
J.Med.Chem., 64, 2021
|
|
1NMR
 
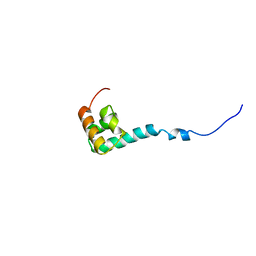 | | Solution Structure of C-terminal Domain from Trypanosoma cruzi Poly(A)-Binding Protein | | Descriptor: | poly(A)-binding protein | | Authors: | Siddiqui, N, Kozlov, G, D'Orso, I, Trempe, J.F, Frasch, A.C.C, Gehring, K. | | Deposit date: | 2003-01-10 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain from poly(A)-binding protein in Trypanosoma cruzi: A vegetal PABC domain
Protein Sci., 12, 2003
|
|
7D5U
 
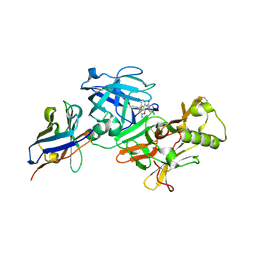 | | BACE2 xaperone complex with N-{3-[(9S)-7-amino-2,2-difluoro-9-(prop-1-yn-1-yl)-6-oxa-8-azaspiro[3.5]non-7-en-9-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 2, N-[3-[(9S)-7-azanyl-2,2-bis(fluoranyl)-9-prop-1-ynyl-6-oxa-8-azaspiro[3.5]non-7-en-9-yl]-4-fluoranyl-phenyl]-5-cyano-pyridine-2-carboxamide, xaperone | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
7D2V
 
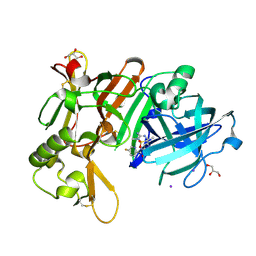 | | Crystal Structure of BACE1 in complex with N-{3-[(5R)-3-amino-2,5-dimethyl-1,1-dioxo-5,6-dihydro-2H-1lambda6,2,4-thiadiazin-5-yl]-4-fluorophenyl}-5-fluoropyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
7D5B
 
 | | BACE2 xaperone complex with N-{3-[(5R)-3-amino-2,5-dimethyl-1,1-dioxo-5,6-dihydro-2H-1lambda6,2,4-thiadiazin-5-yl]-4-fluorophenyl}-5-fluoropyridine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, Beta-secretase 2, CHLORIDE ION, ... | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
6IA3
 
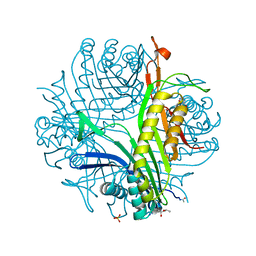 | | urate oxidase under 220 bar (22 MPa) of argon | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-AZAXANTHINE, ARGON, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
1NZ5
 
 | | The Horse heart myoglobin variant K45E/K63E complexed with Manganese | | Descriptor: | MANGANESE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hunter, C.L, Maurus, R, Mauk, M.R, Lee, H, Raven, E.L, Tong, H, Nguyen, N, Smith, S, Brayer, G.D, Mauk, A.G. | | Deposit date: | 2003-02-15 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Introduction and characterization of a functionally linked metal ion binding site at
the exposed heme edge of myoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4UER
 
 | | 40S-eIF1-eIF1A-eIF3-eIF3j translation initiation complex from Lachancea kluyveri | | Descriptor: | 18S RRNA, EIF1, EIF1A, ... | | Authors: | Aylett, C.H.S, Boehringer, D, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-12-18 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | Structure of a Yeast 40S-Eif1-Eif1A-Eif3-Eif3J Initiation Complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UIJ
 
 | | Crystal structure of the BTB domain of KCTD13 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN 1, CHLORIDE ION | | Authors: | Pinkas, D.M, Sanvitale, C.E, Sorell, F.J, Solcan, N, Goubin, S, Canning, P, Williams, E, Chaikuad, A, Dixon Clarke, S.E, Tallant, C, Fonseca, M, Chalk, R, Doutch, J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
4UP1
 
 | |
4UEG
 
 | | Crystal structure of human glycogenin-2 catalytic domain | | Descriptor: | GLYCOGENIN-2, MAGNESIUM ION | | Authors: | Fairhead, M, Strain-Damerell, C, Krojer, T, Froese, D.S, Kopec, J, Nowak, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-12-17 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of Human Glycogenin-2 Catalytic Domain
To be Published
|
|
7DED
 
 | | Mevo lectin complex with mannoheptose (Man7) | | Descriptor: | 1,2-ETHANEDIOL, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, ... | | Authors: | Sivaji, N, Surolia, A, Vijayan, M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.228 Å) | | Cite: | Mevo lectin specificity toward high-mannose structures with terminal alpha Man(1,2) alpha Man residues and its implication to inhibition of the entry of Mycobacterium tuberculosis into macrophages.
Glycobiology, 31, 2021
|
|
