1IUC
 
 | | Fucose-specific lectin from Aleuria aurantia with three ligands | | Descriptor: | Fucose-specific lectin, SULFATE ION, alpha-L-fucopyranose, ... | | Authors: | Fujihashi, M, Peapus, D.H, Kamiya, N, Nagata, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-01 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of Fucose-Specific Lectin from Aleuria aurantia Binding Ligands at Three of Its Five Sugar Recognition Sites
Biochemistry, 42, 2003
|
|
1WJZ
 
 | | Soluiotn structure of J-domain of mouse DnaJ like protein | | Descriptor: | 1700030A21Rik protein | | Authors: | Kobayashi, N, Koshiba, S, Inoue, M, Tochio, N, Tomizawa, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Soluiotn structure of J-domain of mouse DnaJ like protein
To be Published
|
|
7DKD
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Asn-Tyr | | Descriptor: | ASPARAGINE, Dipeptidyl-peptidase, GLYCEROL, ... | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKC
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Tyr-Tyr | | Descriptor: | Dipeptidyl-peptidase, GLYCEROL, TYROSINE | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKE
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Phe-Tyr | | Descriptor: | Dipeptidyl-peptidase, GLYCEROL, PHENYLALANINE, ... | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKB
 
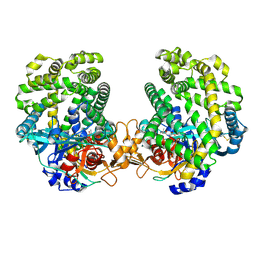 | | Stenotrophomonas maltophilia DPP7 in complex with Val-Tyr | | Descriptor: | Dipeptidyl-peptidase, TYROSINE, VALINE | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
5Y7Z
 
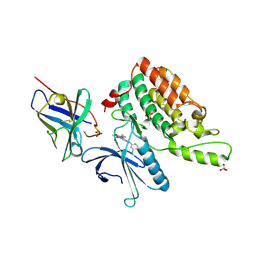 | | Complex structure of cyclin G-associated kinase with gefitinib | | Descriptor: | Cyclin-G-associated kinase, Gefitinib, NANOBODY, ... | | Authors: | Ohbayashi, N, Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural Basis for the Inhibition of Cyclin G-Associated Kinase by Gefitinib.
ChemistryOpen, 7, 2018
|
|
1WYB
 
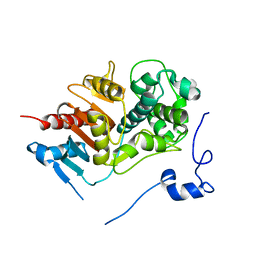 | | Structure of 6-aminohexanoate-dimer hydrolase | | Descriptor: | 6-aminohexanoate-dimer hydrolase | | Authors: | Negoro, S, Ohki, T, Shibata, N, Mizuno, N, Wakitani, Y, Tsurukame, J, Matsumoto, K, Kawamoto, I, Takeo, M, Higuchi, Y. | | Deposit date: | 2005-02-09 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of 6-aminohexanoate-dimer hydrolase
To be Published
|
|
6J3Z
 
 | | Structure of C2S1M1-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
1WWX
 
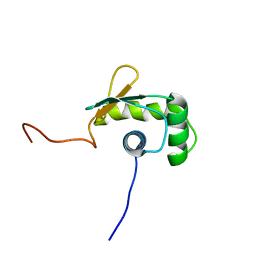 | | Solution structure of the ETS-domain of the Ets domain transcription factor | | Descriptor: | E74-like factor 5 ESE-2b | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ETS-domain of the Ets domain transcription factor
To be Published
|
|
1EYN
 
 | | Structure of mura liganded with the extrinsic fluorescence probe ANS | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, GLYCEROL, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYLTRANSFERASE | | Authors: | Schonbrunn, E, Eschenburg, S, Luger, K, Kabsch, W, Amrhein, N. | | Deposit date: | 2000-05-07 | | Release date: | 2000-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6J3Y
 
 | | Structure of C2S2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
1WFR
 
 | | Solution structure of the conserved hypothetical protein TT1886, possibly sterol carrier protein, from Thermus Thermophilus HB8 | | Descriptor: | Hypothetical Protein TT1886 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the conserved hypothetical protein TT1886, possibly sterol carrier protein, from Thermus Thermophilus HB8
To be Published
|
|
1WQZ
 
 | | Complicated water orientations in the minor groove of B-DNA decamer D(CCATTAATGG)2 observed by neutron diffraction measurements | | Descriptor: | 5'-D(*CP*CP*AP*TP*TP*AP*AP*TP*GP*G)-3' | | Authors: | Arai, S, Chatake, T, Ohhara, T, Kurihara, K, Tanaka, I, Suzuki, N, Fujimoto, Z, Mizuno, H, Niimura, N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (3 Å) | | Cite: | Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements
Nucleic Acids Res., 33, 2005
|
|
1WR6
 
 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | Descriptor: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | Authors: | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-12 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
1IO5
 
 | | HYDROGEN AND HYDRATION OF HEN EGG-WHITE LYSOZYME DETERMINED BY NEUTRON DIFFRACTION | | Descriptor: | LYSOZYME C | | Authors: | Niimura, N, Minezaki, Y, Nonaka, T, Castagna, J.C, Cipriani, F, Hoeghoej, P, Lehmann, M.S, Wilkinson, C. | | Deposit date: | 2001-01-14 | | Release date: | 2001-02-07 | | Last modified: | 2024-10-16 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Neutron Laue diffractometry with an imaging plate provides an effective data collection regime for neutron protein crystallography.
Nat.Struct.Biol., 4, 1997
|
|
5YJO
 
 | |
1IZL
 
 | | Crystal Structure of Photosystem II | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Kamiya, N, Shen, J.-R. | | Deposit date: | 2002-10-04 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-A resolution
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1IS8
 
 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex plus Zn | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IXV
 
 | | Crystal Structure Analysis of homolog of oncoprotein gankyrin, an interactor of Rb and CDK4/6 | | Descriptor: | Probable 26S proteasome regulatory subunit p28 | | Authors: | Padmanabhan, B, Adachi, N, Kataoka, K, Horikoshi, M. | | Deposit date: | 2002-07-09 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the homolog of the oncoprotein gankyrin, an interactor of Rb and CDK4/6
J.BIOL.CHEM., 279, 2004
|
|
7NN6
 
 | |
1WJI
 
 | | Solution Structure of the UBA Domain of Human Tudor Domain Containing Protein 3 | | Descriptor: | Tudor domain containing protein 3 | | Authors: | Kamatari, Y.O, Tochio, N, Nakanishi, T, Miyamoto, K, Li, H, Kobayashi, N, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the UBA Domain of Human Tudor Domain Containing Protein 3
To be Published
|
|
5YE5
 
 | | structure of endo-lysosomal TRPML1 channel inserting into nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, mammalian endo-lysosomal TRPML1 channel | | Authors: | Yang, M, Gao, N. | | Deposit date: | 2017-09-15 | | Release date: | 2017-11-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Cryo-EM structures of the mammalian endo-lysosomal TRPML1 channel elucidate the combined regulation mechanism
Protein Cell, 8, 2017
|
|
1WQY
 
 | | X-RAY structural analysis of B-DNA decamer D(CCATTAATGG)2 crystal grown in D2O solution | | Descriptor: | 5'-D(*CP*CP*AP*TP*TP*AP*AP*TP*GP*G)-3' | | Authors: | Arai, S, Chatake, T, Ohhara, T, Kurihara, K, Tanaka, I, Suzuki, N, Fujimoto, Z, Mizuno, H, Niimura, N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements
Nucleic Acids Res., 33, 2005
|
|
1WWV
 
 | | Solution structure of the SAM domain of human connector enhancer of KSR-like protein CNK1 | | Descriptor: | Connector enhancer of kinase suppressor of ras 1 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM domain of human connector enhancer of KSR-like protein CNK1
To be Published
|
|
