3L84
 
 | | High resolution crystal structure of transketolase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | ACETATE ION, GLYCEROL, Transketolase | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-29 | | Release date: | 2010-02-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | High resolution crystal structure of transketolase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
3LU8
 
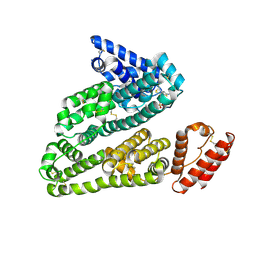 | | Human serum albumin in complex with compound 3 | | Descriptor: | N-[5-(5-{[(2,4-dimethyl-1,3-thiazol-5-yl)sulfonyl]amino}-6-fluoropyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]acetamide, Serum albumin | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
3LD4
 
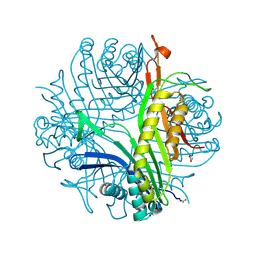 | | Urate oxidase complexed with 8-nitro xanthine | | Descriptor: | 1,2-ETHANEDIOL, 8-nitro-3,7-dihydro-1H-purine-2,6-dione, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N, Gabison, L. | | Deposit date: | 2010-01-12 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Near-atomic resolution structures of urate oxidase complexed with its substrate and analogues: the protonation state of the ligand.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1P4C
 
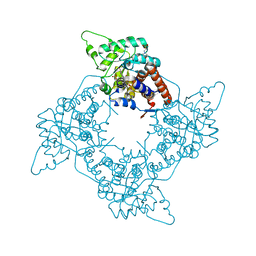 | | High Resolution Structure of Oxidized Active Mutant of (S)-Mandelate Dehydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, L(+)-Mandelate Dehydrogenase, ... | | Authors: | Sukumar, N, Mitra, B, Mathews, F.S. | | Deposit date: | 2003-04-22 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution Structures of an Oxidized and Reduced Flavoprotein: THE WATER SWITCH IN A SOLUBLE FORM OF (S)-MANDELATE DEHYDROGENASE
J.Biol.Chem., 279, 2004
|
|
7B83
 
 | | Structure of SARS-CoV-2 Main Protease bound to pyrithione zinc | | Descriptor: | 3C-like proteinase, 9-oxa-7-thia-1-azonia-8$l^{2}-zincabicyclo[4.3.0]nona-1,3,5-triene, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
3LD6
 
 | | Crystal structure of human lanosterol 14alpha-demethylase (CYP51) in complex with ketoconazole | | Descriptor: | 1-acetyl-4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazine, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Lanosterol 14-alpha demethylase, ... | | Authors: | Strushkevich, N, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of human CYP51 inhibition by antifungal azoles.
J.Mol.Biol., 397, 2010
|
|
1P5B
 
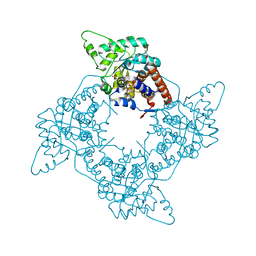 | | High Resolution Structure of Reduced Active Mutant of (S)-Mandelate Dehydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, L(+)-Mandelate Dehydrogenase, ... | | Authors: | Sukumar, N, Mitra, B, Mathews, F.S. | | Deposit date: | 2003-04-25 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution structures of an oxidized and reduced flavoprotein. The water switch in a soluble form of (S)-mandelate dehydrogenase
J.Biol.Chem., 279, 2004
|
|
1OSJ
 
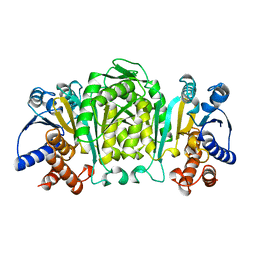 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
1OI8
 
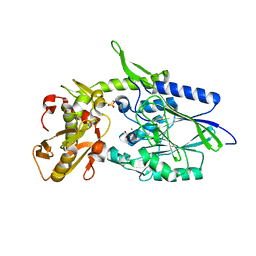 | | 5'-Nucleotidase (E. coli) with an Engineered Disulfide Bridge (P90C, L424C) | | Descriptor: | CARBONATE ION, MANGANESE (II) ION, PROTEIN USHA, ... | | Authors: | Schultz-Heienbrok, R, Maier, T, Straeter, N. | | Deposit date: | 2003-06-10 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trapping a 96 Degree Domain Rotation in Two Distinct Conformations by Engineered Disulfide Bridges
Protein Sci., 13, 2004
|
|
1OIE
 
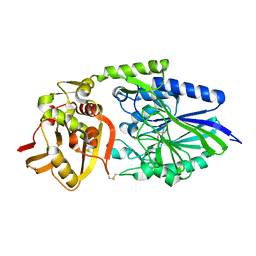 | |
1OSI
 
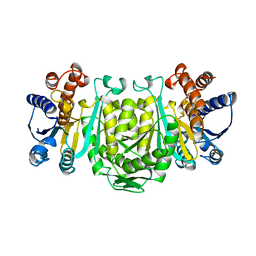 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
4WZM
 
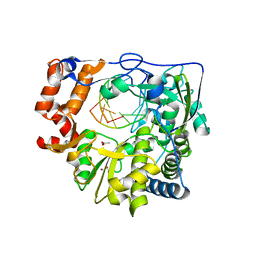 | | Mutant K18E of RNA dependent RNA polymerase from Foot-and-Mouth Disease Virus complexed with RNA | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Verdaguer, N, Ferrer-Orta, C, de la Higuera, N. | | Deposit date: | 2014-11-20 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Multifunctionality of a picornavirus polymerase domain: nuclear localization signal and nucleotide recognition.
J.Virol., 89, 2015
|
|
8DB3
 
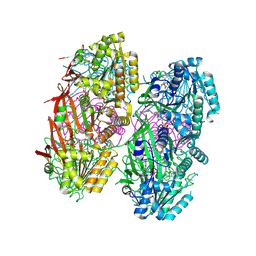 | | Crystal structure of KaiC with truncated C-terminal coiled-coil domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiC | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8DBA
 
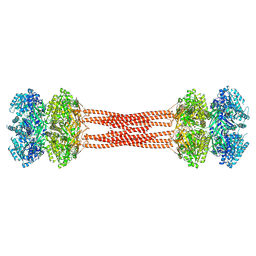 | | Crystal structure of dodecameric KaiC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiC, MAGNESIUM ION | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
5Y4H
 
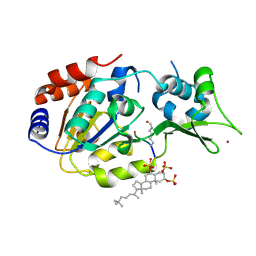 | | Human SIRT3 in complex with halistanol sulfate | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, THr-Arg-Ser-GLY-ALY-VAL-MET-ARG-ARG-LEU-ARG-ARG, ... | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Halistanol sulfates I and J, new SIRT1-3 inhibitory steroid sulfates from a marine sponge of the genus Halichondria
J. Antibiot., 71, 2018
|
|
7O3V
 
 | | Stalk complex structure (TrwJ/VirB5-TrwI/VirB6) from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwI protein, TrwJ protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O3T
 
 | | I-layer structure (TrwF/VirB9NTD, TrwE/VirB10NTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwE protein, TrwF protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O43
 
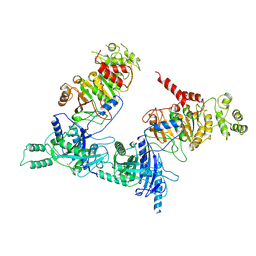 | |
7O42
 
 | | TrwK/VirB4unbound trimer of dimers complex (with Hcp1) from the R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwK protein,Protein hcp1 | | Authors: | Vadakkepat, A.K, Mace, K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O3J
 
 | | O-layer structure (TrwH/VirB7, TrwF/VirB9CTD, TrwE/VirB10CTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwE protein, TrwF protein, TrwH protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-01 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7OIU
 
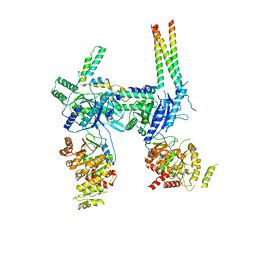 | | Inner Membrane Complex (IMC) protomer structure (TrwM/VirB3, TrwK/VirB4, TrwG/VirB8tails) from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwG protein, TrwK protein, TrwM protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-05-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O41
 
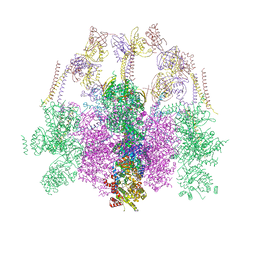 | | Hexameric composite model of the Inner Membrane Complex (IMC) with the Arches from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwG protein, TrwK protein, TrwM protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O4D
 
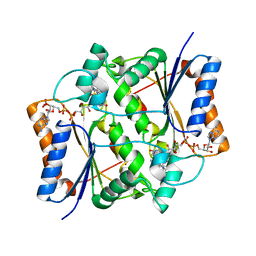 | | QR2 inhibitor from a novel sulfanamide series to tackle age related oxidative stress and cognitive decline | | Descriptor: | 8-methyl-2-(4-methyl-3-piperazin-1-ylsulfonyl-phenyl)imidazo[1,2-a]pyridine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Gould, N.L, Scherer, G.R, Carvalh, S, Shurrush, K, Edry, E, Elkobi, A, David, O, Dym, O, Albeck, S, Peleg, Y, Germain, N, Babaev, I, Sharir, H, Lefker, B, Subramanyam, C, Barr, H, Rosenblum, K. | | Deposit date: | 2021-04-06 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Specific quinone reductase 2 inhibitors reduce metabolic burden and reverse Alzheimer's disease phenotype in mice.
J.Clin.Invest., 133, 2023
|
|
1NAW
 
 | | ENOLPYRUVYL TRANSFERASE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYL-TRANSFERASE | | Authors: | Schoenbrunn, E, Sack, S, Eschenburg, S, Perrakis, A, Krekel, F, Amrhein, N, Mandelkow, E. | | Deposit date: | 1996-07-23 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin.
Structure, 4, 1996
|
|
7OY4
 
 | | VDR complex of a side-chain hydroxylated derivatives of lithocholic acid | | Descriptor: | (3S,6R)-6-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]heptane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2021-06-23 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis and evaluation of side-chain hydroxylated derivatives of lithocholic acid as potent agonists of the vitamin D receptor (VDR).
Bioorg.Chem., 115, 2021
|
|
