6OS4
 
 | | Calmodulin in complex with farnesyl cysteine methyl ester | | Descriptor: | CALCIUM ION, Calmodulin-1, s-farnesyl-l-cysteine methyl ester | | Authors: | Grant, B.M.M, Enomoto, M, Lee, K.Y, Back, S.I, Gebregiworgis, T, Ishiyama, N, Ikura, M, Marshall, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Calmodulin disrupts plasma membrane localization of farnesylated KRAS4b by sequestering its lipid moiety.
Sci.Signal., 13, 2020
|
|
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6CGQ
 
 | | Threonine synthase from Bacillus subtilis ATCC 6633 with PLP and PLP-Ala | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Petronikolou, N, Nair, S.K. | | Deposit date: | 2018-02-20 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Molecular Basis of Bacillus subtilis ATCC 6633 Self-Resistance to the Phosphono-oligopeptide Antibiotic Rhizocticin.
ACS Chem. Biol., 14, 2019
|
|
6Q2L
 
 | |
6CHG
 
 | | Crystal structure of the yeast COMPASS catalytic module | | Descriptor: | H3, Histone-lysine N-methyltransferase, H3 lysine-4 specific, ... | | Authors: | Hsu, P.L, Li, H, Zheng, N. | | Deposit date: | 2018-02-22 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal Structure of the COMPASS H3K4 Methyltransferase Catalytic Module.
Cell, 174, 2018
|
|
6CHV
 
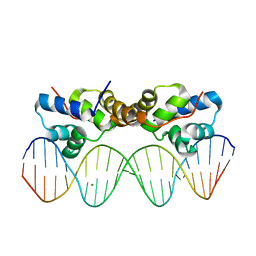 | | Proteus vulgaris HigA antitoxin bound to DNA | | Descriptor: | Antitoxin HigA, MAGNESIUM ION, pHigCryst3, ... | | Authors: | Schureck, M.A, Hoffer, E.D, Onuoha, N, Dunham, C.M. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
1SAC
 
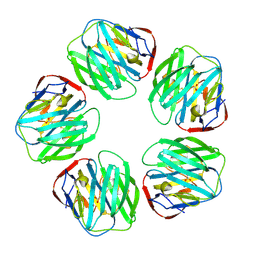 | | THE STRUCTURE OF PENTAMERIC HUMAN SERUM AMYLOID P COMPONENT | | Descriptor: | ACETIC ACID, CALCIUM ION, SERUM AMYLOID P COMPONENT | | Authors: | White, H.E, Emsley, J, O'Hara, B.P, Oliva, G, Srinivasan, N, Tickle, I.J, Blundell, T.L, Pepys, M.B, Wood, S.P. | | Deposit date: | 1994-01-27 | | Release date: | 1994-05-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of pentameric human serum amyloid P component.
Nature, 367, 1994
|
|
1SB9
 
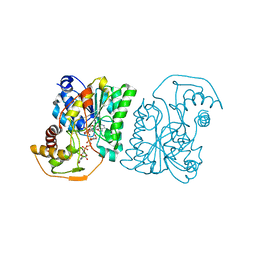 | | Crystal structure of Pseudomonas aeruginosa UDP-N-acetylglucosamine 4-epimerase complexed with UDP-glucose | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCOSE, wbpP | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2004-02-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of WbpP, a Genuine UDP-N-acetylglucosamine 4-Epimerase from Pseudomonas aeruginosa: SUBSTRATE SPECIFICITY IN UDP-HEXOSE 4-EPIMERASES.
J.Biol.Chem., 279, 2004
|
|
6PTZ
 
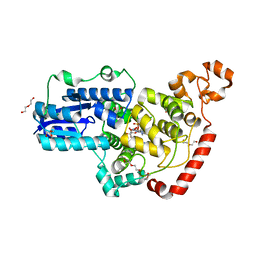 | | Crystal structure of pigeon Cryptochrome 4 mutant Y319D in complex with flavin adenine dinucleotide | | Descriptor: | Cryptochrome-1, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zoltowski, B.D, Chelliah, Y, Wickramaratne, A.C, Jarocha, L, Karki, N, Mouritsen, H, Hore, P.J, Hibbs, R.E, Green, C.B, Takahashi, J.S. | | Deposit date: | 2019-07-16 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Chemical and structural analysis of a photoactive vertebrate cryptochrome from pigeon.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GF1
 
 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | Descriptor: | SULFATE ION, Ubiquitin D | | Authors: | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | Deposit date: | 2018-04-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
1SD6
 
 | | Crystal Structure of Native MecI at 2.65 A | | Descriptor: | Methicillin resistance regulatory protein mecI | | Authors: | Safo, M.K, Zhao, Q, Musayev, F.N, Robinson, H, Scarsdale, N, Archer, G.L. | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
5DFZ
 
 | | Structure of Vps34 complex II from S. cerevisiae. | | Descriptor: | Nanobody binding S. cerevisiae Vps34, Phosphatidylinositol 3-kinase VPS34, Putative N-terminal domain of S. cerevisiae Vps30, ... | | Authors: | Rostislavleva, K, Soler, N, Ohashi, Y, Zhang, L, Williams, R.L. | | Deposit date: | 2015-08-27 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes.
Science, 350, 2015
|
|
6Q3U
 
 | | Gly52Ala mutant of arginine-bound ArgBP from T. maritima | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein, ... | | Authors: | Balasco, N, Smaldone, G, Ruggiero, A, Vitagliano, L. | | Deposit date: | 2018-12-04 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The characterization of Thermotoga maritima Arginine Binding Protein variants demonstrates that minimal local strains have an important impact on protein stability.
Sci Rep, 9, 2019
|
|
6PVK
 
 | | Bacterial 45SRbgA ribosomal particle class A | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Ortega, J, Seffouh, A, Jain, N, Jahagirdar, D, Basu, K, Razi, A, Ni, X, Guarne, A, Britton, R.A. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate.
Nucleic Acids Res., 47, 2019
|
|
6GTB
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with FB211 | | Descriptor: | 1,2-ETHANEDIOL, 17-beta-hydroxysteroid dehydrogenase 14, 3-[6-(3-hydroxyphenyl)pyridin-2-yl]benzoic acid, ... | | Authors: | Bertoletti, N, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2018-06-18 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | X-ray Crystallographic Fragment screening and Hit Optimization
To Be Published
|
|
6VYH
 
 | | Cryo-EM structure of SLC40/ferroportin in complex with Fab | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Ren, Z, Pan, Y, Gao, S, Yan, N, Zhou, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-11 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ion transport and inhibition in ferroportin.
Nat Commun, 11, 2020
|
|
6CO6
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB | | Descriptor: | GLYCEROL, Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6Q71
 
 | | Crystal structure of the alanine racemase Bsu17640 from Bacillus subtilis in the presence of Bis-Tris propane | | Descriptor: | Alanine racemase 2, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bernardo-Garcia, N, Gago, F, Hermoso, J.A. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Cold-induced aldimine bond cleavage by Tris in Bacillus subtilis alanine racemase.
Org.Biomol.Chem., 17, 2019
|
|
5EE5
 
 | | Structure of human ARL1 in complex with the DCB domain of BIG1 | | Descriptor: | ACETATE ION, ADP-ribosylation factor-like protein 1, Brefeldin A-inhibited guanine nucleotide-exchange protein 1, ... | | Authors: | Galindo, A, Soler, N, Munro, S. | | Deposit date: | 2015-10-22 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Structural Insights into Arl1-Mediated Targeting of the Arf-GEF BIG1 to the trans-Golgi.
Cell Rep, 16, 2016
|
|
6Q88
 
 | | RT structure of HEWL at 5 kGy | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Lysozyme C | | Authors: | de la Mora, E, Coquelle, N, Bury, C.S, Rosenthal, M, Garman, E.F, Burghammer, M, Colletier, J.P, Weik, M. | | Deposit date: | 2018-12-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74007058 Å) | | Cite: | Radiation damage and dose limits in serial synchrotron crystallography at cryo- and room temperatures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VZS
 
 | | Engineered TTLL6 mutant bound to gamma-elongation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1BGK
 
 | | SEA ANEMONE TOXIN (BGK) WITH HIGH AFFINITY FOR VOLTAGE DEPENDENT POTASSIUM CHANNEL, NMR, 15 STRUCTURES | | Descriptor: | BGK | | Authors: | Dauplais, M, Lecoq, A, Song, J, Cotton, J, Jamin, N, Gilquin, B, Roumestand, C, Vita, C, Harvey, A, Menez, A. | | Deposit date: | 1996-05-08 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures.
J.Biol.Chem., 272, 1997
|
|
6GY4
 
 | | Crystal structure of the N-terminal KH domain of human BICC1 | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, CHLORIDE ION, Protein bicaudal C homolog 1, ... | | Authors: | Newman, J.A, Katis, V.L, Shrestha, L, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-06-28 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Crystal structure of the N-terminal KH domain of human BICC1
To Be Published
|
|
6BLK
 
 | | Mycobacterial sensor histidine kinase MprB | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Signal transduction histidine-protein kinase/phosphatase mprB | | Authors: | Li, J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2017-11-10 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Mycobacterial sensor histidine kinase MprB
to be published
|
|
6W1R
 
 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.23 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
