9D5W
 
 | | Crystal Structure of the Substrate Binding Domain Protein of the ABC Transporter PBP2_YxeM from Vibrio cholerae | | Descriptor: | Amino acid ABC transporter, periplasmic amino acid-binding portion, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Grimshaw, S, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-14 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Substrate Binding Domain Protein of the ABC Transporter PBP2_YxeM from Vibrio cholerae
To Be Published
|
|
9IYH
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.25 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.25 A resolution.
To Be Published
|
|
9F1D
 
 | | Mammalian quaternary complex of a translating 80S ribosome, NAC, MetAP1 and NatA/E-HYPK | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing.
Nature, 633, 2024
|
|
9D6J
 
 | | Nitrile hydratase BR52K mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Miller, C.G, Holz, R.C, Liu, D, Kaley, N. | | Deposit date: | 2024-08-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Role of second-sphere arginine residues in metal binding and metallocentre assembly in nitrile hydratases.
J Inorg Biochem, 256, 2024
|
|
9IYG
 
 | |
8ZYO
 
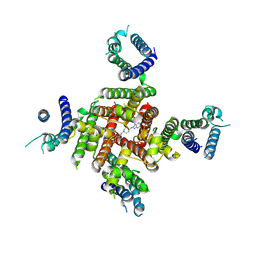 | | Cryo-EM Structure of astemizole-bound hERG Channel | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
9EVV
 
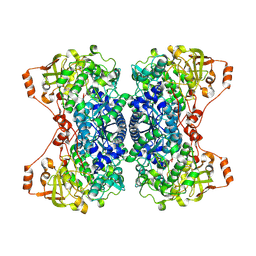 | | His579Leu variant of L-arabinonate dehydratase co-crystallized with 2-oxobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, FE2/S2 (INORGANIC) CLUSTER, L-arabinonate dehydratase, ... | | Authors: | Ren, Y, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2024-04-02 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Unveiling the importance of the C-terminus in the sugar acid dehydratase of the IlvD/EDD superfamily.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
9D6M
 
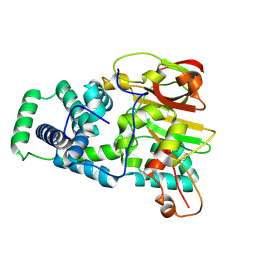 | | Nitrile hydratase BR157K mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Miller, C.G, Holz, R.C, Liu, D, Kaley, N. | | Deposit date: | 2024-08-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Role of second-sphere arginine residues in metal binding and metallocentre assembly in nitrile hydratases.
J Inorg Biochem, 256, 2024
|
|
9CM8
 
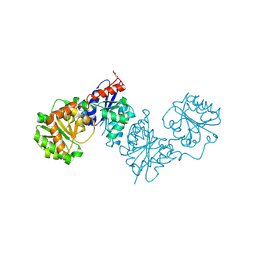 | | UDP-GlcNAc 2-epimerase MnaA of Paenibacillus alvei | | Descriptor: | DI(HYDROXYETHYL)ETHER, UDP-N-acetylglucosamine 2-epimerase (non-hydrolyzing) | | Authors: | Legg, M.S.G, Mateyko, N, Evans, S.V. | | Deposit date: | 2024-07-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into structure and activity of a UDP-GlcNAc 2-epimerase involved in secondary cell wall polymer biosynthesis in Paenibacillus alvei.
Front Mol Biosci, 11, 2024
|
|
8Y58
 
 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with acepromazine. | | Descriptor: | 1-[10-(3-DIMETHYLAMINO-PROPYL)-10H-PHENOTHIAZIN-2-YL]-ETHANONE, E3 ubiquitin-protein ligase TRIM21, FORMIC ACID | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective degradation of multimeric proteins via chemically induced proximity to TRIM21.
To be published
|
|
9EZ3
 
 | | Crystal structure of human CLK3 bound to RN129 | | Descriptor: | 1-(5-~{tert}-butyl-2-quinolin-6-yl-pyrazol-3-yl)-3-[3-(4-morpholin-4-yl-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]urea, Dual specificity protein kinase CLK3, SULFATE ION | | Authors: | Kraemer, A, Raig, N, Knapp, S. | | Deposit date: | 2024-04-10 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human CLK3 bound to RN129
To Be Published
|
|
8Y59
 
 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with (S)-hydroxyl-acepromazine. | | Descriptor: | (1~{S})-1-[10-[3-(dimethylamino)propyl]phenothiazin-2-yl]ethanol, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Selective degradation of multimeric proteins via chemically induced proximity to TRIM21.
To be published
|
|
9BOR
 
 | | IkappaBzeta ankyrin repeat domain:NF-kappaB p50 homodimer complex at 2.0 Angstrom resolution | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, NF-kappa-B inhibitor zeta, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Rogers, W.E, Zhu, N, Huxford, T. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural and biochemical analyses of the nuclear I kappa B zeta protein in complex with the NF-kappa B p50 homodimer.
Genes Dev., 38, 2024
|
|
8Y5B
 
 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with (R)-hydroxyl-acepromazine. | | Descriptor: | (1~{R})-1-[10-[3-(dimethylamino)propyl]phenothiazin-2-yl]ethanol, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Selective degradation of multimeric proteins via chemically induced proximity to TRIM21.
To be published
|
|
5CBU
 
 | | Human Cyclophilin D Complexed with Inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor.
To Be Published
|
|
9BZN
 
 | | High resolution structure of class A Beta-lactamase from Bordetella bronchiseptica RB50 | | Descriptor: | FORMIC ACID, SULFATE ION, class A Beta-lactamase, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-05-24 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | High resolution structure of class A Beta-lactamase from Bordetella bronchiseptica RB50
To Be Published
|
|
9GJ2
 
 | | Crystal structure of human cathepsin S produced in insect cells in complex with ketoamide 13b | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Cathepsin S, ... | | Authors: | Falke, S, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Reinke, P.Y.A, Guenther, S, Turk, D, Meents, A. | | Deposit date: | 2024-08-20 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of human cathepsin S produced in insect cells in complex with ketoamide 13b
To Be Published
|
|
9GP1
 
 | | Jumonji domain-containing protein 2A with crystallization epitope mutatios K330R:A334E | | Descriptor: | Lysine-specific demethylase 4A, NICKEL (II) ION, ZINC ION | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-09-06 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To be published
|
|
9BBM
 
 | |
9D65
 
 | | Nitrile hydratase BR52A mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Miller, C.G, Holz, R.C, Liu, D, Kaley, N. | | Deposit date: | 2024-08-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Role of second-sphere arginine residues in metal binding and metallocentre assembly in nitrile hydratases.
J Inorg Biochem, 256, 2024
|
|
9IYF
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.37 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.37 A resolution.
To Be Published
|
|
9IT8
 
 | | Crystal structure of the ternary complex of lactoperoxidase with nitric oxide and nitrite ion at 1.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maurya, A, Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-19 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Crystal structure of the ternary complex of lactoperoxidase with nitric oxide and nitrite ion at 1.95 A resolution
To Be Published
|
|
8ZYQ
 
 | | Cryo-EM Structure of pimozide-bound hERG Channel | | Descriptor: | 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
9F1B
 
 | | Mammalian ternary complex of a translating 80S ribosome, NAC and NatA/E | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing.
Nature, 633, 2024
|
|
9CX4
 
 | |
