6ZZC
 
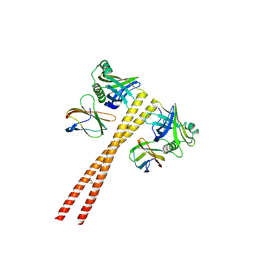 | | MB_CRS6-1 bound to CrSAS-6_6HR | | 分子名称: | Centriole protein, DODECAETHYLENE GLYCOL, MB_CrS6-1 | | 著者 | Hatzopoulos, G.N, Kukenshoner, T, Banterle, N, Favez, T, Fluckiger, I, Hantschel, O, Gonczy, P. | | 登録日 | 2020-08-04 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly.
Nat Commun, 12, 2021
|
|
6P6D
 
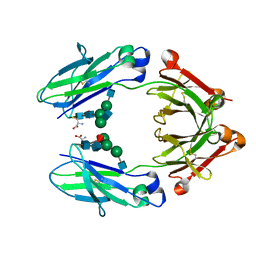 | | HUMAN IGG1 FC FRAGMENT, C239 INSERTION MUTANT | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, IgG1 Fc fragment | | 著者 | Gallagher, D.T, Dimasi, N, McCullough, C. | | 登録日 | 2019-06-03 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structure and Dynamics of a Site-Specific Labeled Fc Fragment with Altered Effector Functions.
Pharmaceutics, 11, 2019
|
|
5YGY
 
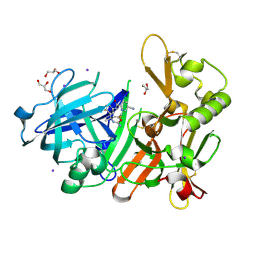 | | Crystal Structure of BACE1 in complex with (S)-N-(3-(2-amino-6-(fluoromethyl)-4 -methyl-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-cyanopicolinamide | | 分子名称: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | 著者 | Fuchino, K, Mitsuoka, Y, Masui, M, Kurose, N, Yoshida, S, Komano, K, Yamamoto, T, Ogawa, M, Unemura, C, Hosono, M, Ito, H, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Miyajima, H, Hiroyama, S, Koyabu, K, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | 登録日 | 2017-09-27 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Rational Design of Novel 1,3-Oxazine Based beta-Secretase (BACE1) Inhibitors: Incorporation of a Double Bond To Reduce P-gp Efflux Leading to Robust A beta Reduction in the Brain
J. Med. Chem., 61, 2018
|
|
5Y5J
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the presence of NADH (resting state) | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5YIH
 
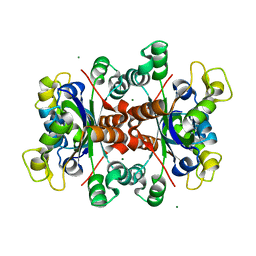 | | Crystal structure of tetrameric Nucleoside diphosphate kinase at 1.98 A resolution from Acinetobacter baumannii | | 分子名称: | MAGNESIUM ION, Nucleoside diphosphate kinase | | 著者 | Bairagya, H.R, Sikarwar, J, Iqbal, N, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2017-10-04 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structure of tetrameric Nucleoside diphosphate kinase at 1.98 A resolution from Acinetobacter baumannii
To Be Published
|
|
1SIO
 
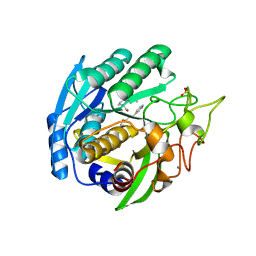 | | Structure of Kumamolisin-As complexed with a covalently-bound inhibitor, AcIPF | | 分子名称: | Ace-ILE-PRO-PHL peptide inhibitor, CALCIUM ION, SULFATE ION, ... | | 著者 | Li, M, Wlodawer, A, Gustchina, A, Tsuruoka, N, Ashida, M, Minakata, H, Oyama, H, Oda, K, Nishino, T, Nakayama, T. | | 登録日 | 2004-03-01 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystallographic and biochemical investigations of kumamolisin-As, a serine-carboxyl peptidase with collagenase activity
J.Biol.Chem., 279, 2004
|
|
2LCI
 
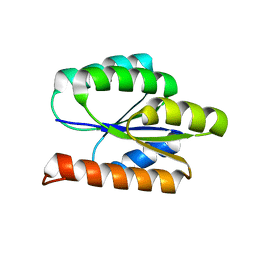 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, P-LOOP NTPASE FOLD, Northeast Structural Genomics Consortium Target OR36 (CASD Target) | | 分子名称: | Protein OR36 | | 著者 | Liu, G, Koga, N, Koga, R, Xiao, R, Janjua, H, Ciccosanti, C, Lee, H, Acton, T.B, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-04-29 | | 公開日 | 2011-06-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Northeast Structural Genomics Consortium Target OR36
To be Published
|
|
6PBC
 
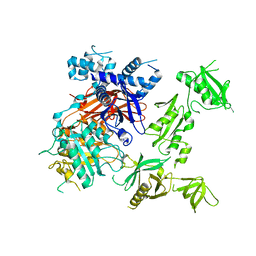 | | Structural basis for the activation of PLC-gamma isozymes by phosphorylation and cancer-associated mutations | | 分子名称: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma,1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, CALCIUM ION, SODIUM ION | | 著者 | Hajicek, N, Sondek, J. | | 登録日 | 2019-06-13 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Structural basis for the activation of PLC-gamma isozymes by phosphorylation and cancer-associated mutations.
Elife, 8, 2019
|
|
5YCU
 
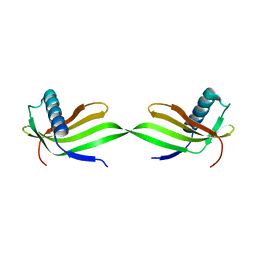 | | Domain swapped dimer of engineered hairpin loop1 mutant in Single-chain Monellin | | 分子名称: | Single chain monellin | | 著者 | Surana, P, Nandwani, N, Udgaonkar, J.B, Gosavi, S, Das, R. | | 登録日 | 2017-09-08 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
7A4P
 
 | | Structure of small high-light grown Chlorella ohadii photosystem I | | 分子名称: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | 著者 | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | 登録日 | 2020-08-20 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
7JRK
 
 | |
6ZZY
 
 | | Structure of high-light grown Chlorella ohadii photosystem I | | 分子名称: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ... | | 著者 | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | 登録日 | 2020-08-05 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
6ZZX
 
 | | Structure of low-light grown Chlorella ohadii Photosystem I | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | 著者 | Caspy, I, Nelson, N, Nechushtai, R, Neumann, E, Shkolnisky, Y. | | 登録日 | 2020-08-05 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
7JRD
 
 | |
1SJQ
 
 | | NMR Structure of RRM1 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | | 分子名称: | Polypyrimidine tract-binding protein 1 | | 著者 | Simpson, P.J, Monie, T.P, Szendroi, A, Davydova, N, Tyzack, J.K, Conte, M.R, Read, C.M, Cary, P.D, Svergun, D.I, Konarev, P.V, Petoukhov, M.V, Curry, S, Matthews, S.J. | | 登録日 | 2004-03-04 | | 公開日 | 2004-09-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and RNA Interactions of the N-Terminal RRM Domains of PTB
Structure, 12, 2004
|
|
5YH4
 
 | |
2LSX
 
 | | Solution structure of a mini i-motif | | 分子名称: | DNA (5'-D(P*TP*CP*GP*TP*TP*TP*CP*GP*TP*T)-3') | | 著者 | Escaja, N, Viladoms, J, Garavis, M, Villasante, A, Pedroso, E, Gonzalez, C. | | 登録日 | 2012-05-09 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A minimal i-motif stabilized by minor groove G:T:G:T tetrads.
Nucleic Acids Res., 40, 2012
|
|
1SH3
 
 | | Crystal Structure of Norwalk Virus Polymerase (MgSO4 crystal form) | | 分子名称: | MAGNESIUM ION, RNA Polymerase | | 著者 | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | 登録日 | 2004-02-24 | | 公開日 | 2004-03-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
6OZ7
 
 | | Putative oxidoreductase from Escherichia coli str. K-12 | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, TRIETHYLENE GLYCOL, ... | | 著者 | Osipiuk, J, Skarina, T, Mesa, N, Endres, M, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-05-15 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Putative oxidoreductase from Escherichia coli str. K-12
to be published
|
|
6PPR
 
 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP and DNA | | 分子名称: | ATP-dependent DNA helicase (UvrD/REP), DNA (70-MER), IRON/SULFUR CLUSTER, ... | | 著者 | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | 登録日 | 2019-07-08 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P00
 
 | |
7K38
 
 | |
5XXW
 
 | | GDP-microtubule complexed with KIF5C in ATP state | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Morikawa, M, Shigematsu, H, Nitta, R, Hirokawa, N. | | 登録日 | 2017-07-05 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport
J. Cell Biol., 217, 2018
|
|
2L79
 
 | |
1SM5
 
 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | 分子名称: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*(BRU)P*TP*AP*AP*TP*(BRU)P*CP*G)-3' | | 著者 | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.-S, Kang, C. | | 登録日 | 2004-03-08 | | 公開日 | 2004-05-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of a DNA Decamer Containing a Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
