4V4O
 
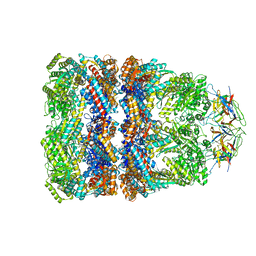 | | Crystal Structure of the Chaperonin Complex Cpn60/Cpn10/(ADP)7 from Thermus Thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Shimamura, T, Koike-Takeshita, A, Yokoyama, K, Masui, R, Murai, N, Yoshida, M, Taguchi, H, Iwata, S. | | Deposit date: | 2004-05-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity
STRUCTURE, 12, 2004
|
|
4V7Q
 
 | | Atomic model of an infectious rotavirus particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Core scaffold protein, ... | | Authors: | Settembre, E.C, Chen, J.Z, Dormitzer, P.R, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2010-05-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic model of an infectious rotavirus particle.
Embo J., 30, 2011
|
|
4V8D
 
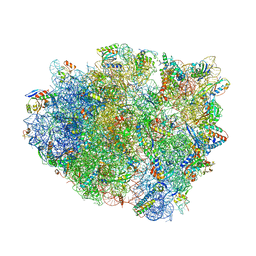 | | Structure analysis of ribosomal decoding (cognate tRNA-tyr complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4UY4
 
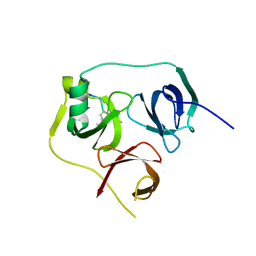 | | 1.86 A structure of human Spindlin-4 protein in complex with histone H3K4me3 peptide | | Descriptor: | GLYCEROL, HISTONE H3K4ME3, SPINDLIN-4 | | Authors: | Talon, R, Gileadi, C, Johansson, C, Burgess-Brown, N, Shrestha, L, von Delft, F, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | 1.86 A Structure of Human Spindlin-4 Protein in Complex with Histone H3K4Me3 Peptide
To be Published
|
|
4URN
 
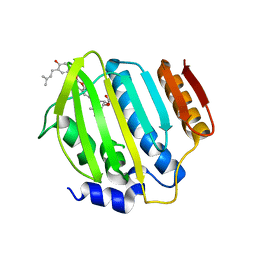 | | Crystal Structure of Staph ParE 24kDa in complex with Novobiocin | | Descriptor: | DNA TOPOISOMERASE IV, B SUBUNIT, NOVOBIOCIN | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4URJ
 
 | | Crystal structure of human BJ-TSA-9 | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN FAM83A | | Authors: | Pinkas, D.M, Sanvitale, C, Wang, D, Krojer, T, Kopec, J, Chaikuad, A, Dixon Clarke, S, Berridge, G, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal Structure of Human Bj-Tsa-9
To be Published
|
|
4USS
 
 | | Populus trichocarpa glutathione transferase X1-1 (GHR1), complexed with glutathione | | Descriptor: | GLUTATHIONE, GLUTATHIONYL HYDROQUINONE REDUCTASE, PHOSPHATE ION | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Dumaracay, S, Favier, F, Didierjean, C, Saul, F, Haouz, A, Morel-Rouhier, M, Gelhaye, E, Rouhier, N, Hecker, A. | | Deposit date: | 2014-07-13 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutathionyl-Hydroquinone Reductases from Poplar are Plastidial Proteins that Deglutathionylate Both Reduced and Oxidized Glutathionylated Quinones.
FEBS Lett., 589, 2015
|
|
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V6Y
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in classic pre-translocation state (pre1a) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4V7C
 
 | | Structure of the Ribosome with Elongation Factor G Trapped in the Pre-Translocation State (pre-translocation 70S*tRNA structure) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Brilot, A.F, Korostelev, A.A, Ermolenko, D.N, Grigorieff, N. | | Deposit date: | 2013-11-20 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the ribosome with elongation factor G trapped in the pretranslocation state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V7A
 
 | | E. coli 70S-fMetVal-tRNAVal post-translocation complex (post4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4V8E
 
 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-tyr complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4WBR
 
 | |
4V59
 
 | | Crystal structure of fatty acid synthase complexed with nadp+ from thermomyces lanuginosus at 3.1 angstrom resolution. | | Descriptor: | FATTY ACID SYNTHASE ALPHA SUBUNITS, FATTY ACID SYNTHASE BETA SUBUNITS, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Jenni, S, Leibundgut, M, Boehringer, D, Frick, C, Mikolasek, B, Ban, N. | | Deposit date: | 2007-03-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Fungal Fatty Acid Synthase and Implications for Iterative Substrate Shuttling
Science, 316, 2007
|
|
4V77
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate post-translocation state (post2b) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4V8L
 
 | |
4UQI
 
 | | AP2 controls clathrin polymerization with a membrane-activated switch | | Descriptor: | AP-2 COMPLEX SUBUNIT ALPHA-2, AP-2 COMPLEX SUBUNIT BETA, AP-2 COMPLEX SUBUNIT MU, ... | | Authors: | Kelly, B.T, Graham, S.C, Liska, N, Dannhauser, P.N, Hoening, S, Ungewickell, E.J, Owen, D.J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-07-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Clathrin Adaptors. Ap2 Controls Clathrin Polymerization with a Membrane-Activated Switch.
Science, 345, 2014
|
|
4V47
 
 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the EF-G.GTP state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
4UWF
 
 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-9-[3,5-bis(fluoranyl)phenyl]-2-morpholin-4-yl-8-(trifluoromethyl)-7,8-dihydro-6H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3 | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxobutyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
4UWY
 
 | | FGFR1 Apo structure | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FIBROBLAST GROWTH FACTOR RECEPTOR 1 | | Authors: | Thiyagarajan, N, Bunney, T, Katan, M. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | The Effect of Mutations on Drug Sensitivity and Kinase Activity of Fibroblast Growth Factor Receptors: A Combined Experimental and Theoretical Study
Ebiomedicine, 2, 2015
|
|
4UXZ
 
 | | Structure of delta7-DgkA-syn in 7.9 MAG to 2.18 angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, ACETATE ION, ... | | Authors: | Li, D, Howe, N, Caffrey, M. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Ternary Structure Reveals Mechanism of a Membrane Diacylglycerol Kinase.
Nat.Commun., 6, 2015
|
|
4V58
 
 | | Crystal structure of fatty acid synthase from thermomyces lanuginosus at 3.1 angstrom resolution. | | Descriptor: | FATTY ACID SYNTHASE ALPHA SUBUNITS, FATTY ACID SYNTHASE BETA SUBUNITS, FLAVIN MONONUCLEOTIDE | | Authors: | Jenni, S, Leibundgut, M, Boehringer, D, Frick, C, Mikolasek, B, Ban, N. | | Deposit date: | 2007-03-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Fungal Fatty Acid Synthase and Implications for Iterative Substrate Shuttling
Science, 316, 2007
|
|
6Z5N
 
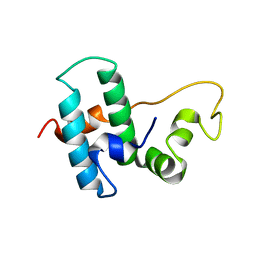 | | DnaJB1 JD-GF | | Descriptor: | DnaJ homolog subfamily B member 1 | | Authors: | Avraham-Abayev, M, London, N, Rosenzweig, R. | | Deposit date: | 2020-05-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | HSP40 proteins use class-specific regulation to drive HSP70 functional diversity.
Nature, 587, 2020
|
|
6ZSH
 
 | |
6ZSI
 
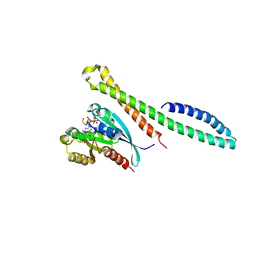 | | The mechanism of activation of the actin binding protein EHBP1 by Rab8 family members. | | Descriptor: | DI(HYDROXYETHYL)ETHER, EH domain-binding protein 1, MAGNESIUM ION, ... | | Authors: | Rai, A, Bleimling, N, Vetter, I.R, Goody, R.S. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | The mechanism of activation of the actin binding protein EHBP1 by Rab8 family members.
Nat Commun, 11, 2020
|
|
