1KFA
 
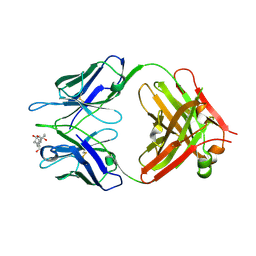 | | Crystal structure of Fab fragment complexed with gibberellin A4 | | Descriptor: | GIBBERELLIN A4, monoclonal antibody heavy chain, monoclonal antibody light chain | | Authors: | Murata, T, Fushinobu, S, Nakajima, M, Asami, O, Sassa, T, Wakagi, T, Yamaguchi, I. | | Deposit date: | 2001-11-20 | | Release date: | 2002-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the liganded anti-gibberellin A(4) antibody 4-B8(8)/E9 Fab fragment.
Biochem.Biophys.Res.Commun., 293, 2002
|
|
1YT6
 
 | | NMR structure of peptide SD | | Descriptor: | peptide SD | | Authors: | Murata, T, Hemmi, H, Nakamura, S, Shimizu, K, Suzuki, Y, Yamaguchi, I. | | Deposit date: | 2005-02-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure, epitope mapping, and docking simulation of a gibberellin mimic peptide as a peptidyl mimotope for a hydrophobic ligand.
Febs J., 272, 2005
|
|
2DB4
 
 | | Crystal structure of rotor ring with DCCD of the V- ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DICYCLOHEXYLUREA, SODIUM ION, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Shirouzu, M, Walker, J.E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2BL2
 
 | | The membrane rotor of the V-type ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, SODIUM ION, UNDECYL-MALTOSIDE, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2005-02-25 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Rotor of the Vacuolar-Type Na- ATPase from Enterococcus Hirae
Science, 308, 2005
|
|
2CYD
 
 | | Crystal structure of Lithium bound rotor ring of the V-ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, LITHIUM ION, UNDECYL-MALTOSIDE, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Shirouzu, M, Walker, J.E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2006-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Lithium bound rotor ring of the V-ATPase from Enterococcus hirae
To be Published
|
|
8ZYO
 
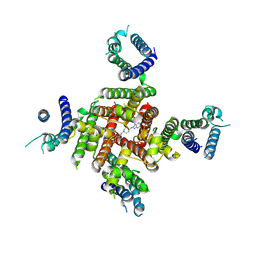 | | Cryo-EM Structure of astemizole-bound hERG Channel | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 32, 2024
|
|
8ZYQ
 
 | | Cryo-EM Structure of pimozide-bound hERG Channel | | Descriptor: | 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 32, 2024
|
|
8ZYP
 
 | | Cryo-EM Structure of E-4031-bound hERG Channel | | Descriptor: | Potassium voltage-gated channel subfamily H member 2, ~{N}-[4-[1-[2-(6-methylpyridin-2-yl)ethyl]piperidin-4-yl]carbonylphenyl]methanesulfonamide | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 32, 2024
|
|
8ZYN
 
 | | Cryo-EM Structure of inhibitor-free hERG Channel | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 32, 2024
|
|
8WCI
 
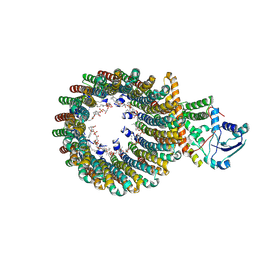 | | Cryo-EM structure of the inhibitor-bound Vo complex from Enterococcus hirae | | Descriptor: | CARDIOLIPIN, N,N-dimethyl-4-(5-methyl-1H-benzimidazol-2-yl)aniline, SODIUM ION, ... | | Authors: | Suzuki, K, Mikuriya, S, Adachi, N, Kawasaki, M, Senda, T, Moriya, T, Murata, T. | | Deposit date: | 2023-09-12 | | Release date: | 2024-10-09 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Na + -V-ATPase inhibitor curbs VRE growth and unveils Na + pathway structure.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8Y6H
 
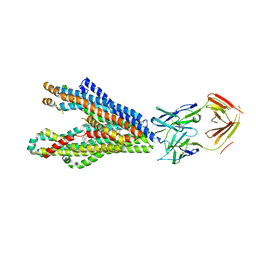 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in LMNG detergent | | Descriptor: | ATP-dependent translocase ABCB1,mNeonGreen, UIC2 Fab heavy chain, UIC2 Fab light chain, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
8Y6I
 
 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in nanodisc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent translocase ABCB1,mNeonGreen, CHOLESTEROL, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
3O0R
 
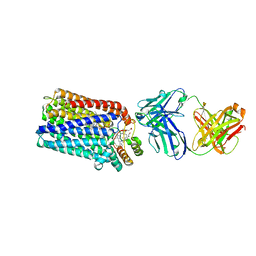 | | Crystal structure of nitric oxide reductase from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, FE (III) ION, HEME C, ... | | Authors: | Hino, T, Matsumoto, Y, Nagano, S, Sugimoto, H, Fukumori, Y, Murata, T, Iwata, S, Shiro, Y. | | Deposit date: | 2010-07-20 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of biological N2O generation by bacterial nitric oxide reductase
Science, 330, 2010
|
|
8XX8
 
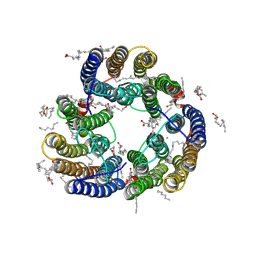 | | Structure of Glycilhalorhodopsin from Salinarimonas soli | | Descriptor: | CHLORIDE ION, EICOSANE, OLEIC ACID, ... | | Authors: | Suzuki, K, Ishizuka, T, Konno, M, Inoue, K, Murata, T. | | Deposit date: | 2024-01-17 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Light-driven anion-pumping rhodopsin with unique cytoplasmic anion-release mechanism.
J.Biol.Chem., 300, 2024
|
|
5KNC
 
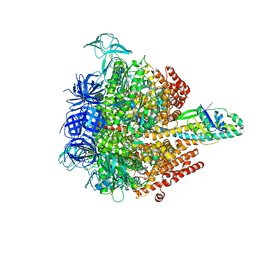 | | Crystal structure of the 3 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.015 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5KNB
 
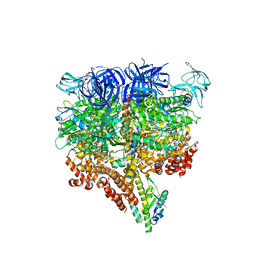 | | Crystal structure of the 2 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5KND
 
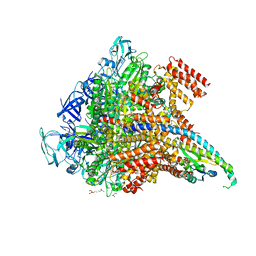 | | Crystal structure of the Pi-bound V1 complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
8XW5
 
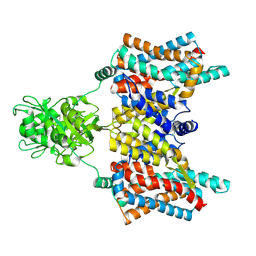 | | Cryo-EM structure of the aspartate:alanine antiporter AspT mutant L60C | | Descriptor: | Aspartate/alanine antiporter | | Authors: | Nanatani, K, Kanno, R, Kawabata, T, Watanabe, S, Hidaka, M, Yamanaka, T, Toda, K, Fujiki, T, Kunii, K, Miyamoto, A, Chiba, F, Ogasawara, S, Murata, T, Humbel, B.M, Inaba, K, Mitsuoka, K, Guan, L, Abe, K, Yamamoto, M, Koshiba, S. | | Deposit date: | 2024-01-16 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure and molecular mechanism of the aspartate:alanine antiporter AspT from Tetragenococcus halophilus
To Be Published
|
|
8Y1X
 
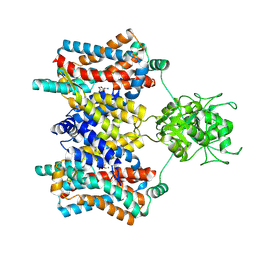 | | Cryo-EM structure of the aspartate:alanine antiporter AspT WT | | Descriptor: | ASPARTIC ACID, Aspartate/alanine antiporter | | Authors: | Nanatani, K, Kanno, R, Kawabata, T, Watanabe, S, Hidaka, M, Yamanaka, T, Toda, K, Fujiki, T, Kunii, K, Miyamoto, A, Chiba, F, Ogasawara, S, Murata, T, Humbel, B.M, Inaba, K, Mitsuoka, K, Guan, L, Abe, K, Yamamoto, M, Koshiba, S. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM structure and molecular mechanism of the aspartate:alanine antiporter AspT from Tetragenococcus halophilus
To Be Published
|
|
7CN0
 
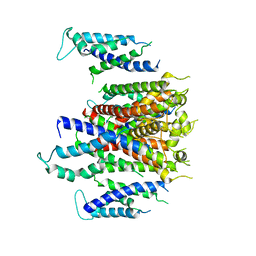 | | Cryo-EM structure of K+-bound hERG channel | | Descriptor: | POTASSIUM ION, potassium channel 1 | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
7CN1
 
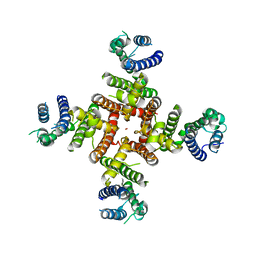 | | Cryo-EM structure of K+-bound hERG channel in the presence of astemizole | | Descriptor: | POTASSIUM ION, potassium channel | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
6KFQ
 
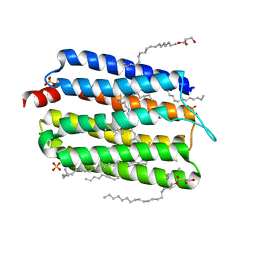 | | Crystal structure of thermophilic rhodopsin from Rubrobacter xylanophilus | | Descriptor: | RETINAL, Rhodopsin, SULFATE ION, ... | | Authors: | Suzuki, K, Akiyama, T, Hayashi, T, Yasuda, S, Kanehara, K, Kojima, K, Tanabe, M, Kato, R, Senda, T, Sudo, Y, Kinoshita, M, Murata, T. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Does a Microbial Rhodopsin RxR Realize Its Exceptionally High Thermostability with the Proton-Pumping Function Being Retained?
J.Phys.Chem.B, 124, 2020
|
|
6IIE
 
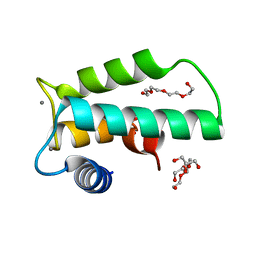 | | Crystal structure of human diacylglycerol kinase alpha EF-hand domains bound to Ca2+ | | Descriptor: | CALCIUM ION, Diacylglycerol kinase alpha, GLYCEROL, ... | | Authors: | Takahashi, D, Suzuki, K, Sakamoto, T, Iwamoto, T, Murata, T, Sakane, F. | | Deposit date: | 2018-10-04 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Crystal structure and calcium-induced conformational changes of diacylglycerol kinase alpha EF-hand domains.
Protein Sci., 28, 2019
|
|
5A16
 
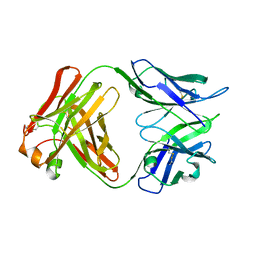 | | Crystal structure of Fab4201 raised against Human Erythrocyte Anion Exchanger 1 | | Descriptor: | FAB4201 HEAVY CHAIN | | Authors: | Arakawa, T, Kobayashi-Yugiri, T, Alguel, Y, Weyand, S, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Ikeda-Suno, C, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Anion Exchanger Domain of Human Erythrocyte Band 3
Science, 350, 2015
|
|
4YZF
 
 | | Crystal structure of the anion exchanger domain of human erythrocyte Band 3 | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Band 3 anion transport protein, FAB fragment of Immunoglobulin (IgG) molecule | | Authors: | Alguel, Y, Arakawa, T, Yugiri, T.K, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Suno, C.I, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A.D, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-03-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the anion exchanger domain of human erythrocyte band 3.
Science, 350, 2015
|
|
