1FQY
 
 | | STRUCTURE OF AQUAPORIN-1 AT 3.8 A RESOLUTION BY ELECTRON CRYSTALLOGRAPHY | | 分子名称: | AQUAPORIN-1 | | 著者 | Murata, K, Mitsuoka, K, Hirai, T, Walz, T, Agre, P, Heymann, J.B, Engel, A, Fujiyoshi, Y. | | 登録日 | 2000-09-07 | | 公開日 | 2000-10-18 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (3.8 Å) | | 主引用文献 | Structural determinants of water permeation through aquaporin-1.
Nature, 407, 2000
|
|
4W7H
 
 | | Crystal Structure of DEH Reductase A1-R Mutant | | 分子名称: | Carbonyl reductase | | 著者 | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | 登録日 | 2014-08-22 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
3K8V
 
 | |
4W7I
 
 | | Crystal structure of DEH reductase A1-R' mutant | | 分子名称: | 4-deoxy-L-erythro-5-hexoseulose uronate reductase A1-R' | | 著者 | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | 登録日 | 2014-08-22 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
2AHF
 
 | |
6S2C
 
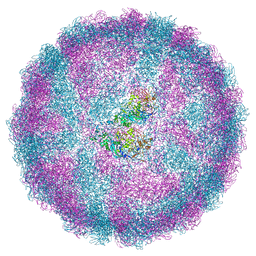 | | Acquired functional capsid structures in metazoan totivirus-like dsRNA virus. | | 分子名称: | Capsid protein | | 著者 | Okamoto, K, Larsson, S.D.D, Maia, R.N.C.F, Murata, K, Hajdu, J, Iwasaki, K, Miyazaki, N. | | 登録日 | 2019-06-20 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Acquired Functional Capsid Structures in Metazoan Totivirus-like dsRNA Virus.
Structure, 28, 2020
|
|
4MMH
 
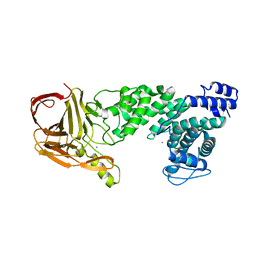 | | Crystal structure of heparan sulfate lyase HepC from Pedobacter heparinus | | 分子名称: | CALCIUM ION, Heparinase III protein | | 著者 | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2013-09-09 | | 公開日 | 2014-01-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
4MMI
 
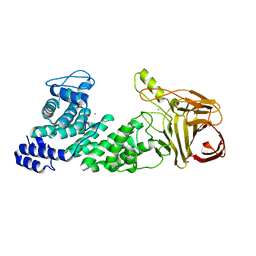 | | Crystal structure of heparan sulfate lyase HepC mutant from Pedobacter heparinus | | 分子名称: | CALCIUM ION, Heparinase III protein | | 著者 | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2013-09-09 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
1AT9
 
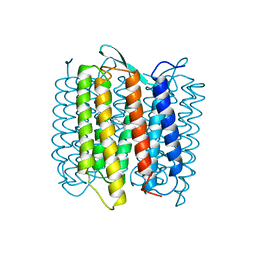 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | | 分子名称: | BACTERIORHODOPSIN, RETINAL | | 著者 | Kimura, Y, Vassylyev, D.G, Miyazawa, A, Kidera, A, Matsushima, M, Mitsuoka, K, Murata, K, Hirai, T, Fujiyoshi, Y. | | 登録日 | 1997-08-20 | | 公開日 | 1998-09-16 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | 主引用文献 | Surface of bacteriorhodopsin revealed by high-resolution electron crystallography.
Nature, 389, 1997
|
|
7YJL
 
 | |
4XIG
 
 | | Crystal structure of bacterial alginate ABC transporter determined through humid air and glue-coating method | | 分子名称: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, AlgM1, AlgM2, ... | | 著者 | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2015-01-07 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.402 Å) | | 主引用文献 | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
4XTC
 
 | | Crystal structure of bacterial alginate ABC transporter in complex with alginate pentasaccharide-bound periplasmic protein | | 分子名称: | AlgM1, AlgM2, AlgQ2, ... | | 著者 | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2015-01-23 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
5SW1
 
 | | Thaumatin Structure at pH 6.0 | | 分子名称: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Thaumatin I | | 著者 | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | 登録日 | 2016-08-08 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Thaumatin Structure at pH 6.0
To Be Published
|
|
5SW2
 
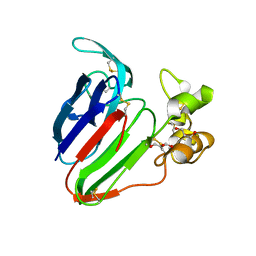 | | Thaumatin Structure at pH 6.0, orthorhombic type1 | | 分子名称: | GLYCEROL, Thaumatin I | | 著者 | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | 登録日 | 2016-08-08 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Thaumatin Structure at pH 6.0, orthorhombic type1
To Be Published
|
|
2OKX
 
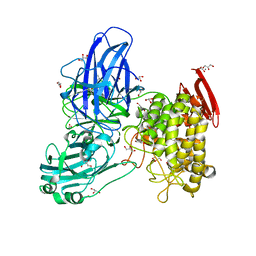 | | Crystal structure of GH78 family rhamnosidase of Bacillus SP. GL1 AT 1.9 A | | 分子名称: | CALCIUM ION, GLYCEROL, Rhamnosidase B | | 著者 | Cui, Z, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2007-01-17 | | 公開日 | 2007-11-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Glycoside Hydrolase Family 78 alpha-L-Rhamnosidase from Bacillus sp. GL1
J.Mol.Biol., 374, 2007
|
|
1VAV
 
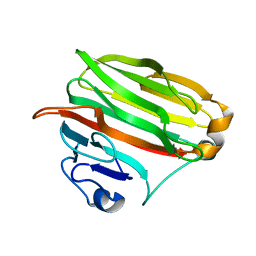 | | Crystal structure of alginate lyase PA1167 from Pseudomonas aeruginosa at 2.0 A resolution | | 分子名称: | Alginate lyase PA1167 | | 著者 | Yamasaki, M, Moriwaki, S, Miyake, O, Hashimoto, W, Murata, K, Mikami, B. | | 登録日 | 2004-02-19 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7: a novel alginate lyase with a beta-sandwich fold.
J.Biol.Chem., 279, 2004
|
|
7X30
 
 | | Capsid structure of Staphylococcus jumbo bacteriophage S6 | | 分子名称: | Hoc-like protein ORF90, Major structural protein ORF12 | | 著者 | Koibuchi, W, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | 登録日 | 2022-02-27 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Capsid structure of Staphylococcus jumbo bacteriophage S6
To Be Published
|
|
3IM0
 
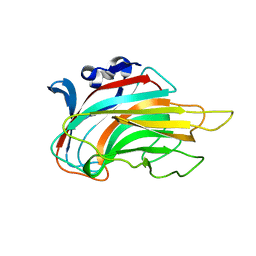 | | Crystal structure of Chlorella virus vAL-1 soaked in 200mM D-glucuronic acid, 10% PEG-3350, and 200mM glycine-NaOH (pH 10.0) | | 分子名称: | VAL-1, beta-D-glucopyranuronic acid | | 著者 | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2009-08-08 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
3GNE
 
 | | Crystal structure of alginate lyase vAL-1 from Chlorella virus | | 分子名称: | CITRATE ANION, GLYCEROL, VAL-1 | | 著者 | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2009-03-17 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
4F13
 
 | | Alginate lyase A1-III Y246F complexed with tetrasaccharide | | 分子名称: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | 著者 | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | 登録日 | 2012-05-06 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.208 Å) | | 主引用文献 | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4E1Y
 
 | | Alginate lyase A1-III H192A apo form | | 分子名称: | Alginate lyase | | 著者 | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | 登録日 | 2012-03-07 | | 公開日 | 2012-04-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F10
 
 | | Alginate lyase A1-III H192A complexed with tetrasaccharide | | 分子名称: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | 著者 | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | 登録日 | 2012-05-05 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2RGK
 
 | | Functional annotation of Escherichia coli yihS-encoded protein | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Uncharacterized sugar isomerase yihS | | 著者 | Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2007-10-03 | | 公開日 | 2008-08-26 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of YihS in complex with D-mannose: structural annotation of Escherichia coli and Salmonella enterica yihS-encoded proteins to an aldose-ketose isomerase
J.Mol.Biol., 377, 2008
|
|
7YCA
 
 | | Cryo-EM structure of the PSI-LHCI-Lhcp supercomplex from Ostreococcus tauri | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Shan, J, Sheng, X, Ishii, A, Watanabe, A, Song, C, Murata, K, Minagawa, J, Liu, Z. | | 登録日 | 2022-07-01 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers.
Elife, 12, 2023
|
|
1J1N
 
 | | Structure Analysis of AlgQ2, A Macromolecule(Alginate)-Binding Periplasmic Protein Of Sphingomonas Sp. A1., Complexed with an Alginate Tetrasaccharide | | 分子名称: | AlgQ2, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | 著者 | Momma, K, Mikami, B, Mishima, Y, Hashimoto, W, Murata, K. | | 登録日 | 2002-12-11 | | 公開日 | 2003-06-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of AlgQ2, a macromolecule (alginate)-binding protein of Sphingomonas sp. A1, complexed with an alginate tetrasaccharide at 1.6-A resolution
J.BIOL.CHEM., 278, 2003
|
|
