1XX1
 
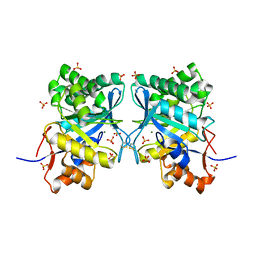 | | Structural basis for ion-coordination and the catalytic mechanism of sphingomyelinases D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Murakami, M.T, Tambourgi, D.V, Arni, R.K. | | Deposit date: | 2004-11-03 | | Release date: | 2005-01-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for metal ion coordination and the catalytic mechanism of sphingomyelinases d
J.Biol.Chem., 280, 2005
|
|
1ZLB
 
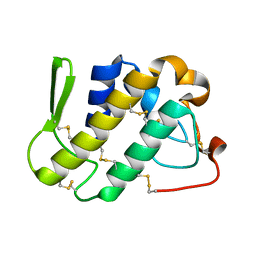 | | Crystal structure of catalytically-active phospholipase A2 in the absence of calcium | | Descriptor: | hypotensive phospholipase A2 | | Authors: | Murakami, M.T, Cintra, A.C, Gabdoulkhakov, A, Genov, N, Betzel, C, Arni, R.K. | | Deposit date: | 2005-05-05 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Insights into metal ion binding in phospholipases A(2): ultra high-resolution crystal structures of an acidic phospholipase A(2) in the Ca(2+) free and bound states.
Biochimie, 88, 2006
|
|
1ZL7
 
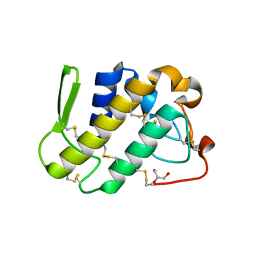 | | Crystal structure of catalytically-active phospholipase A2 with bound calcium | | Descriptor: | CALCIUM ION, GLYCEROL, hypotensive phospholipase A2 | | Authors: | Murakami, M.T, Cintra, A.C, Gabdoulkhakov, A, Genov, N, Betzel, C, Arni, R.K. | | Deposit date: | 2005-05-05 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into metal ion binding in phospholipases A(2): ultra high-resolution crystal structures of an acidic phospholipase A(2) in the Ca(2+) free and bound states.
Biochimie, 88, 2006
|
|
1XXN
 
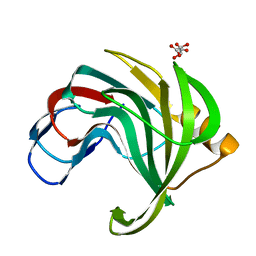 | | Crystal structure of a mesophilic xylanase A from Bacillus subtilis 1A1 | | Descriptor: | Endo-1,4-beta-xylanase A, S,R MESO-TARTARIC ACID | | Authors: | Murakami, M.T, Ruller, R, Ward, R.J, Arni, R.K. | | Deposit date: | 2004-11-07 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Correlation of temperature induced conformation change with optimum catalytic activity in the recombinant G/11 xylanase A from Bacillus subtilis strain 168 (1A1).
Febs Lett., 579, 2005
|
|
1Y4L
 
 | | Crystal structure of Bothrops asper myotoxin II complexed with the anti-trypanosomal drug suramin | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Murakami, M.T, Arruda, E.Z, Melo, P.A, Martinez, A.B, Calil-Elias, S, Tomaz, M.A, Lomonte, B, Gutierrez, J.M, Arni, R.K. | | Deposit date: | 2004-12-01 | | Release date: | 2005-06-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Myotoxic Activity of Bothrops asper Myotoxin II by the Anti-trypanosomal Drug Suramin.
J.Mol.Biol., 350, 2005
|
|
2AIP
 
 | |
2AIQ
 
 | |
2PH4
 
 | | Crystal structure of a novel Arg49 phospholipase A2 homologue from Zhaoermia mangshanensis venom | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, Zhaoermiatoxin | | Authors: | Murakami, M.T, Kuch, U, Mebs, D, Arni, R.K. | | Deposit date: | 2007-04-10 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a novel myotoxic Arg49 phospholipase A(2) homolog (zhaoermiatoxin) from Zhaoermia mangshanensis snake venom: Insights into Arg49 coordination and the role of Lys122 in the polarization of the C-terminus.
Toxicon, 51, 2008
|
|
2F9R
 
 | | Crystal structure of the inactive state of the Smase I, a sphingomyelinase D from Loxosceles laeta venom | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Sphingomyelinase D 1 | | Authors: | Murakami, M.T, Gabdoulkhakov, A, Fernandes-Pedrosa, M.F, Betzel, C, Tambourgi, D.V, Arni, R.K. | | Deposit date: | 2005-12-06 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for metal ion coordination and the catalytic mechanism of sphingomyelinases D.
J.Biol.Chem., 280, 2005
|
|
2H9E
 
 | | Crystal Structure of FXa/selectide/NAPC2 ternary complex | | Descriptor: | ACETATE ION, Anti-coagulant protein C2, Coagulation factor X heavy chain, ... | | Authors: | Murakami, M.T, Geiger, G, Tulinsky, A, Arni, R.K. | | Deposit date: | 2006-06-09 | | Release date: | 2007-02-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intermolecular Interactions and Characterization of the Novel Factor Xa Exosite Involved in Macromolecular Recognition and Inhibition: Crystal Structure of Human Gla-domainless Factor Xa Complexed with the Anticoagulant Protein NAPc2 from the Hematophagous Nematode Ancylostoma caninum.
J.Mol.Biol., 366, 2007
|
|
2H8U
 
 | |
2H8I
 
 | |
1UMR
 
 | | Crystal structure of the platelet activator convulxin, a disulfide linked a4b4 cyclic tetramer from the venom of Crotalus durissus terrificus | | Descriptor: | CONVULXIN ALPHA, CONVULXIN BETA | | Authors: | Murakami, M.T, Zela, S.P, Gava, L.M, Michelan-Duarte, S, Cintra, A.C.O, Arni, R.K. | | Deposit date: | 2003-08-28 | | Release date: | 2003-11-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Platelet Activator Convulxin, a Disulfide Linked A4B4 Cyclic Tetramer from the Venom of Crotalus Durissus Terrificus
Biochem.Biophys.Res.Commun., 310, 2003
|
|
1UMV
 
 | | Crystal structure of an acidic, non-myotoxic phospholipase A2 from the venom of Bothrops jararacussu | | Descriptor: | CALCIUM ION, HYPOTENSIVE PHOSPHOLIPASE A2 | | Authors: | Murakami, M.T, Watanabe, L, Cintra, A.C.O, Arni, R.K. | | Deposit date: | 2003-08-28 | | Release date: | 2003-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of an Acidic Platelet Aggregation Inhibitor and Hypotensive Phospholipase A(2) in the Monomeric and Dimeric States: Insights Into its Oligomeric State
Biochem.Biophys.Res.Commun., 323, 2004
|
|
4E2B
 
 | | High resolution crystal structure of the old yellow enzyme from Trypanosoma cruzi | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Murakami, M.T, Rodrigues, N.C, Gava, L.M, Canduri, F, Oliva, G, Barbosa, L.R.S, Borgers, J.C. | | Deposit date: | 2012-03-08 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | High resolution crystal structure and in solution studies of the old yellow enzyme from Trypanosoma cruzi: Insights into oligomerization, enzyme dynamics and specificity
To be Published
|
|
4E2D
 
 | | Structure of the old yellow enzyme from Trypanosoma cruzi | | Descriptor: | DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, Old Yellow Protein | | Authors: | Murakami, M.T, Rodrigues, N.C, Gava, L.M, Canduri, F, Oliva, G, Barbosa, L.R.S, Borgers, J.C. | | Deposit date: | 2012-03-08 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | High resolution crystal structure and in solution studies of the Old Yellow Enzyme from Trypanosoma cruzi: Insights into oligomerization, enzyme dynamics and specificity
To be Published
|
|
4GSO
 
 | | structure of Jararacussin-I | | Descriptor: | Thrombin-like enzyme BjussuSP-1 | | Authors: | Ullah, A, Souza, T.C.A.B, Zanphorlin, L.M, Mariutti, R, Sanata, S.V, Murakami, M.T, Arni, R.K. | | Deposit date: | 2012-08-28 | | Release date: | 2012-12-12 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Jararacussin-I: The highly negatively charged catalytic interface contributes to macromolecular selectivity in snake venom thrombin-like enzymes.
Protein Sci., 22, 2013
|
|
4W7V
 
 | |
4W85
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, beta-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4W8A
 
 | | Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | Exo-xyloglucanase, GLYCEROL, SULFATE ION | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4W84
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
8D89
 
 | | Crystal structure of a novel GH5 enzyme retrieved from capybara gut metagenome | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Martins, M.P, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2022-06-08 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Glycoside hydrolase subfamily GH5_57 features a highly redesigned catalytic interface to process complex hetero-beta-mannans.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7OVA
 
 | | Crystal structure of an AA9 LPMO | | Descriptor: | (2S)-2-hydroxybutanedioic acid, COPPER (II) ION, Endoglucanase, ... | | Authors: | Males, A, Correa, T.L.R, Murakami, M.T, Walton, P.H, Davies, G.J. | | Deposit date: | 2021-06-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of an AA9 LPMO
To Be Published
|
|
3KVE
 
 | | Structure of native L-amino acid oxidase from Vipera ammodytes ammodytes: stabilization of the quaternary structure by divalent ions and structural changes in the dynamic active site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase, ... | | Authors: | Gergiova, D, Murakami, M.T, Perbandt, M, Arni, R.K, Betzel, C. | | Deposit date: | 2009-11-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of native L-amino acid oxidase from Vipera ammodytes ammodytes: stabilization of the quaternary structure by divalent ions and structural changes in the dynamic active site
To be Published
|
|
6UB3
 
 | | Crystal structure of a GH128 (subgroup IV) endo-beta-1,3-glucanase from Lentinula edodes (LeGH128_IV) with laminaribiose at the surface-binding site | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Santos, C.R, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
