8EZ8
 
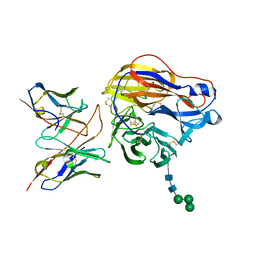 | | Structure of 3C08 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 3C08, Light chain of influenza virus neuraminidase antibody 3C08, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase.
Immunity, 56, 2023
|
|
8EZ7
 
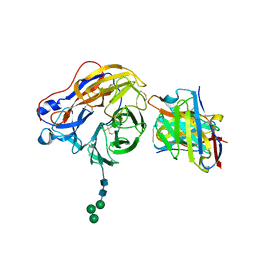 | | Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 1F04, Light chain of influenza virus neuraminidase antibody 1F04, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase.
Immunity, 56, 2023
|
|
8EZ3
 
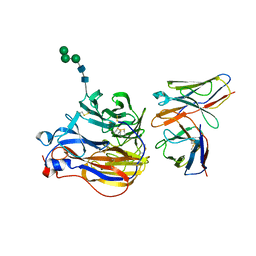 | | Structure of 3A10 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 3A10, Light chain of influenza virus neuraminidase antibody 3A10, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase.
Immunity, 56, 2023
|
|
3IZ3
 
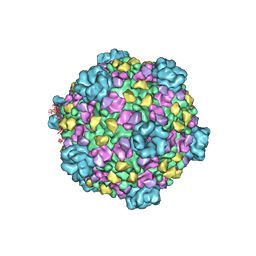 | | CryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Structural protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Cheng, L, Sun, J, Zhang, K, Mou, Z, Huang, X, Ji, G, Sun, F, Zhang, J, Zhu, P. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4JW2
 
 | | Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep) | | Descriptor: | 1,2-ETHANEDIOL, A3 artificial protein, bA3-2: binder of A3 protein | | Authors: | Guellouz, A, Valerio-Lepiniec, M, Urvoas, A, Chevrel, A, Graille, M, Fourati-Kammoun, Z, Desmadril, M, van Tilbeurgh, H, Minard, P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selection of Specific Protein Binders for Pre-Defined Targets from an Optimized Library of Artificial Helicoidal Repeat Proteins (alphaRep).
Plos One, 8, 2013
|
|
4JW3
 
 | | Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep) | | Descriptor: | Alpha-helical artificial proteins, Neocarzinostatin | | Authors: | Guellouz, A, Valerio-Lepiniec, M, Urvoas, A, Chevrel, A, Graille, M, Fourati-Kammoun, Z, Desmadril, M, van Tilbeurgh, H, Minard, P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selection of Specific Protein Binders for Pre-Defined Targets from an Optimized Library of Artificial Helicoidal Repeat Proteins (alphaRep).
Plos One, 8, 2013
|
|
4OJJ
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P212121) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
4OGP
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
2AUV
 
 | | Solution Structure of HndAc : A Thioredoxin-like [2Fe-2S] Ferredoxin Involved in the NADP-reducing Hydrogenase Complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, potential NAD-reducing hydrogenase subunit | | Authors: | Nouailler, M, Morelli, X, Bornet, O, Chetrit, B, Dermoun, Z, Guerlesquin, F. | | Deposit date: | 2005-08-29 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HndAc: a thioredoxin-like domain involved in the NADP-reducing hydrogenase complex
Protein Sci., 15, 2006
|
|
