1J1I
 
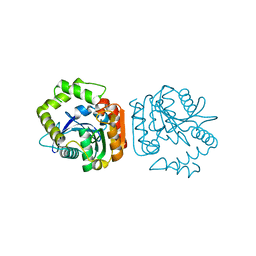 | | Crystal structure of a His-tagged Serine Hydrolase Involved in the Carbazole Degradation (CarC enzyme) | | Descriptor: | meta cleavage compound hydrolase | | Authors: | Habe, H, Morii, K, Fushinobu, S, Nam, J.W, Ayabe, Y, Yoshida, T, Wakagi, T, Yamane, H, Nojiri, H, Omori, T. | | Deposit date: | 2002-12-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a histidine-tagged serine hydrolase involved in the carbazole degradation (CarC enzyme).
Biochem.Biophys.Res.Commun., 303, 2003
|
|
1B90
 
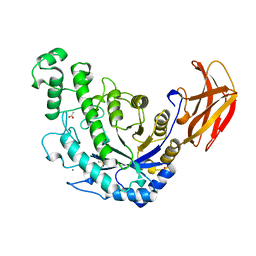 | | BACILLUS CEREUS BETA-AMYLASE APO FORM | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
1B9Z
 
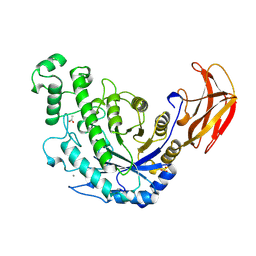 | | BACILLUS CEREUS BETA-AMYLASE COMPLEXED WITH MALTOSE | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
5DKZ
 
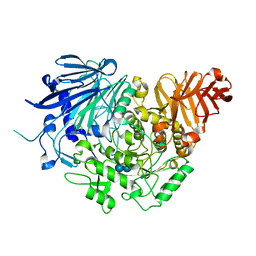 | | Crystal structure of glucosidase II alpha subunit (alpha3-Glc2-bound from) | | Descriptor: | Alpha glucosidase-like protein, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
5DKX
 
 | | Crystal structure of glucosidase II alpha subunit (Tris-bound from) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha glucosidase-like protein, CHLORIDE ION | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
5DL0
 
 | | Crystal structure of glucosidase II alpha subunit (Glc1Man2-bound from) | | Descriptor: | Alpha glucosidase-like protein, alpha-D-glucopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
5DKY
 
 | | Crystal structure of glucosidase II alpha subunit (DNJ-bound from) | | Descriptor: | 1-DEOXYNOJIRIMYCIN, Alpha glucosidase-like protein | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
8EL2
 
 | | SARS-CoV-2 RBD bound to neutralizing antibody Fab ICO-hu23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ICO-hu23 Heavy Chain, Fab ICO-hu23 Light Chain, ... | | Authors: | Besaw, J.E, Kuo, A, Morizumi, T, Ernst, O.P. | | Deposit date: | 2022-09-22 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Broadly neutralizing humanized SARS-CoV-2 antibody binds to a conserved epitope on Spike and provides antiviral protection through inhalation-based delivery in non-human primates.
Plos Pathog., 19, 2023
|
|
8ELJ
 
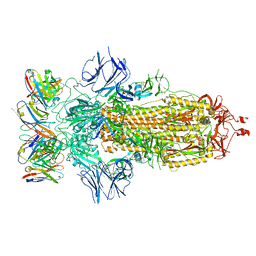 | | SARS-CoV-2 spike glycoprotein in complex with the ICO-hu23 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ICO-hu23 antibody Fab heavy chain, ... | | Authors: | Yee, A.W, Morizumi, T, Kim, K, Kuo, A, Ernst, O.P. | | Deposit date: | 2022-09-24 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing humanized SARS-CoV-2 antibody binds to a conserved epitope on Spike and provides antiviral protection through inhalation-based delivery in non-human primates.
Plos Pathog., 19, 2023
|
|
5H18
 
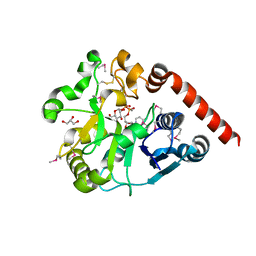 | | Crystal structure of catalytic domain of UGGT (UDP-glucose-bound form) from Thermomyces dupontii | | Descriptor: | CALCIUM ION, GLYCEROL, UGGT, ... | | Authors: | Satoh, T, Zhu, T, Toshimori, T, Kamikubo, H, Uchihashi, T, Kato, K. | | Deposit date: | 2016-10-08 | | Release date: | 2017-09-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Visualisation of a flexible modular structure of the ER folding-sensor enzyme UGGT.
Sci Rep, 7, 2017
|
|
5JQP
 
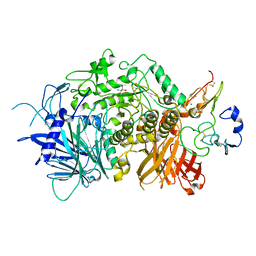 | | Crystal structure of ER glucosidase II heterodimeric complex consisting of catalytic subunit and the binding domain of regulatory subunit | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha glucosidase-like protein, CALCIUM ION, ... | | Authors: | Satoh, T, Toshimori, T, Noda, M, Uchiyama, S, Kato, K. | | Deposit date: | 2016-05-05 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction mode between catalytic and regulatory subunits in glucosidase II involved in ER glycoprotein quality control.
Protein Sci., 25, 2016
|
|
8Y6H
 
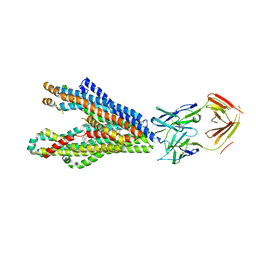 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in LMNG detergent | | Descriptor: | ATP-dependent translocase ABCB1,mNeonGreen, UIC2 Fab heavy chain, UIC2 Fab light chain, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
8Y6I
 
 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in nanodisc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent translocase ABCB1,mNeonGreen, CHOLESTEROL, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
1BJ3
 
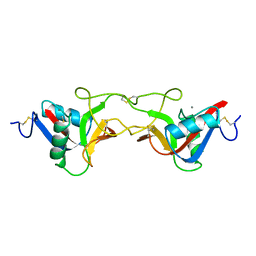 | | CRYSTAL STRUCTURE OF COAGULATION FACTOR IX-BINDING PROTEIN (IX-BP) FROM VENOM OF HABU SNAKE WITH A HETERODIMER OF C-TYPE LECTIN DOMAINS | | Descriptor: | CALCIUM ION, PROTEIN (COAGULATION FACTOR IX-BINDING PROTEIN A), PROTEIN (COAGULATION FACTOR IX-BINDING PROTEIN B) | | Authors: | Mizuno, H, Fujimoto, Z, Koizumi, M, Kano, H, Atoda, H, Morita, T. | | Deposit date: | 1998-07-02 | | Release date: | 1999-08-16 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of coagulation factor IX-binding protein from habu snake venom at 2.6 A: implication of central loop swapping based on deletion in the linker region.
J.Mol.Biol., 289, 1999
|
|
6WP8
 
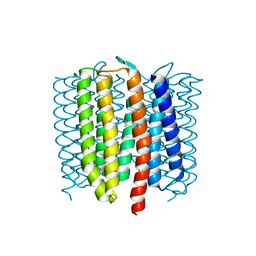 | | Proton-pumping mutant of Mastigocladopsis repens rhodopsin chloride pump | | Descriptor: | Proton-pumping rhodopsin chloride pump, RETINAL, octyl beta-D-glucopyranoside | | Authors: | Besaw, J.E, Ernst, O.P, Ou, W, Morizumi, T. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of a chloride-pumping microbial rhodopsin and its proton-pumping mutant illuminate proton transfer determinants.
J.Biol.Chem., 295, 2020
|
|
6XL3
 
 | | Mastigocladopsis repens rhodopsin chloride pump | | Descriptor: | CHLORIDE ION, DECANE, Mastigocladopsis repens rhodopsin chloride pump, ... | | Authors: | Besaw, J.E, Ernst, O.P, Ou, W, Morizumi, T. | | Deposit date: | 2020-06-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The crystal structures of a chloride-pumping microbial rhodopsin and its proton-pumping mutant illuminate proton transfer determinants.
J.Biol.Chem., 295, 2020
|
|
8T1T
 
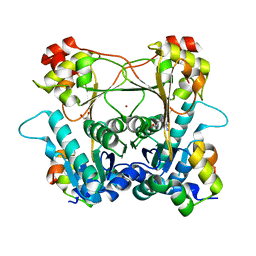 | | Structure of the alpha-N-methyltransferase (SonM) and RiPP precursor (SonA with QSY deletion) heteromeric complex (bound to SAM) | | Descriptor: | Alpha-N-methyltransferase (SonM), RiPP precursor (SonA), S-ADENOSYLMETHIONINE, ... | | Authors: | Crone, K.K, Jomori, T, Miller, F.S, Gralnick, J, Elias, M, Freeman, M.F. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | RiPP enzyme heterocomplex structure-guided discovery of a bacterial borosin alpha- N -methylated peptide natural product.
Rsc Chem Biol, 4, 2023
|
|
8T1S
 
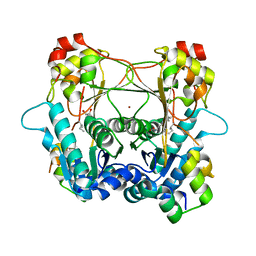 | | Structure of the alpha-N-methyltransferase (SonM) and RiPP precursor (SonA with QSY deletion) heteromeric complex (bound to SAH) | | Descriptor: | Alpha-N-methyltransferase, Extradiol ring-cleavage dioxygenase LigAB LigA subunit domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Crone, K.K, Jomori, T, Miller, F.S, Gralnick, J, Elias, M, Freeman, M.F. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RiPP enzyme heterocomplex structure-guided discovery of a bacterial borosin alpha- N -methylated peptide natural product.
Rsc Chem Biol, 4, 2023
|
|
6NWE
 
 | |
8HW1
 
 | | Far-red light-harvesting complex of Antarctic alga Prasiola crispa | | Descriptor: | (1S)-4-[(1E,3Z,5E,7E,9E,11E,13E,15E,17E)-3-(hydroxymethyl)-7,12,16-trimethyl-18-[(1R,4S)-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]-3,5,5-trimethyl-cyclohex-3-en-1-ol, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, CHLOROPHYLL A, ... | | Authors: | Kosugi, M, Kawasaki, M, Shibata, Y, Moriya, T, Adachi, N, Senda, T. | | Deposit date: | 2022-12-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Uphill energy transfer mechanism for photosynthesis in an Antarctic alga.
Nat Commun, 14, 2023
|
|
8IDQ
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus with xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
8IDP
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
6NWD
 
 | | X-ray Crystallographic structure of Gloeobacter rhodopsin | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DECANE, DODECANE, ... | | Authors: | Ernst, O.P, Morizumi, T, Ou, W.L. | | Deposit date: | 2019-02-06 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Crystallographic Structure and Oligomerization of Gloeobacter Rhodopsin.
Sci Rep, 9, 2019
|
|
6M3X
 
 | | Cryo-EM structure of sulfur oxygenase reductase from Sulfurisphaera tokodaii | | Descriptor: | FE (III) ION, Sulfur oxygenase/reductase | | Authors: | Sato, Y, Adachi, N, Moriya, T, Arakawa, T, Kawasaki, M, Yamada, C, Senda, T, Fushinobu, S. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Crystallographic and cryogenic electron microscopic structures and enzymatic characterization of sulfur oxygenase reductase fromSulfurisphaera tokodaii.
J Struct Biol X, 4, 2020
|
|
8JOR
 
 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type A crystal | | Descriptor: | Acyltransferase, PENTAETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
