2DT5
 
 | | Crystal Structure of TTHA1657 (AT-rich DNA-binding protein) from Thermus thermophilus HB8 | | Descriptor: | AT-rich DNA-binding protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nakamura, A, Sosa, A, Komori, H, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-11 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of TTHA1657 (AT-rich DNA-binding protein; p25) from Thermus thermophilus HB8 at 2.16 A resolution
Proteins, 66, 2007
|
|
1XER
 
 | | STRUCTURE OF FERREDOXIN | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, ZINC ION | | Authors: | Fujii, T, Hata, Y, Moriyama, H, Wakagi, T, Tanaka, N, Oshima, T. | | Deposit date: | 1996-08-28 | | Release date: | 1997-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding centre in thermoacidophilic archaeal ferredoxins.
Nat.Struct.Biol., 3, 1996
|
|
2YXW
 
 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
2CMM
 
 | | STRUCTURAL ANALYSIS OF THE MYOGLOBIN RECONSTITUTED WITH IRON PORPHINE | | Descriptor: | CYANIDE ION, MYOGLOBIN, PORPHYRIN FE(III) | | Authors: | Sato, T, Tanaka, N, Moriyama, H, Igarashi, N, Neya, S, Funasaki, N, Iizuka, T, Shiro, Y. | | Deposit date: | 1993-12-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the myoglobin reconstituted with iron porphine.
J.Biol.Chem., 268, 1993
|
|
2YXV
 
 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
7CIO
 
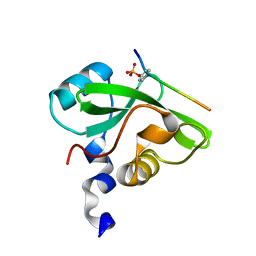 | | Molecular interactions of cytoplasmic region of CTLA-4 with SH2 domains of PI3-kinase | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Phosphatidylinositol 3-kinase regulatory subunit alpha | | Authors: | Iiyama, M, Numoto, N, Ogawa, S, Kuroda, M, Morii, H, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular interactions of the CTLA-4 cytoplasmic region with the phosphoinositide 3-kinase SH2 domains.
Mol.Immunol., 131, 2021
|
|
3B0L
 
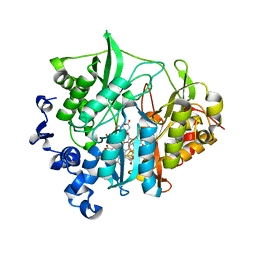 | | M175G mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B8G
 
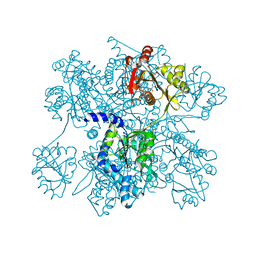 | | Crysta structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and N-acetyl-glutamate | | Descriptor: | COENZYME A, N-ACETYL-L-GLUTAMATE, Putative acetylglutamate synthase | | Authors: | Shi, D, Sagar, V, Jin, Z, Yu, X, Caldovic, L, Morizono, H, Allewell, N.M, Tuchman, M. | | Deposit date: | 2007-11-01 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of N-acetyl-L-glutamate synthase from Neisseria gonorrhoeae provides insights into mechanisms of catalysis and regulation.
J.Biol.Chem., 283, 2008
|
|
3B0M
 
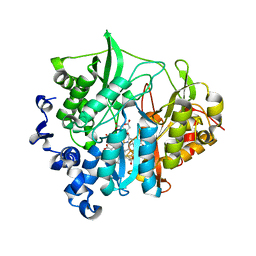 | | M175K mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B0N
 
 | | Q448K mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B0G
 
 | | Assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
7WKF
 
 | | Antimicrobial peptide-LaIT2 | | Descriptor: | Beta-KTx-like peptide LaIT2 | | Authors: | Tamura, M, Morita, H, Ohki, S. | | Deposit date: | 2022-01-09 | | Release date: | 2023-04-05 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of LaIT2, an antimicrobial and insecticidal peptide from Liocheles australasiae.
Toxicon, 214, 2022
|
|
1VGJ
 
 | | Crystal structure of 2'-5' RNA ligase from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH0099 | | Authors: | Yao, M, Morita, H, Okada, A, Tanaka, I. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of Pyrococcus horikoshii 2'-5' RNA ligase at 1.94 A resolution reveals a possible open form with a wider active-site cleft
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
3X39
 
 | | Domain-swapped dimer of Pseudomonas aeruginosa cytochrome c551 | | Descriptor: | Cytochrome c-551, HEME C | | Authors: | Nagao, S, Ueda, M, Osuka, H, Komori, H, Kamikubo, H, Kataoka, M, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Domain-Swapped Dimer of Pseudomonas aeruginosa Cytochrome c551: Structural Insights into Domain Swapping of Cytochrome c Family Proteins
Plos One, 10, 2015
|
|
5AUR
 
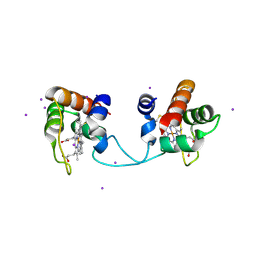 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at N-terminal region | | Descriptor: | Cytochrome c-552, HEME C, IODIDE ION | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Kamikubo, H, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
3VYM
 
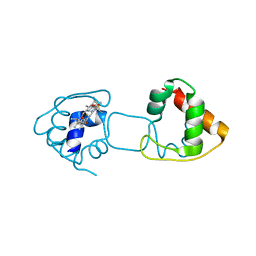 | | Dimeric Hydrogenobacter thermophilus cytochrome c552 | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Hayashi, Y, Nagao, S, Osuka, H, Komori, H, Higuchi, Y, Hirota, S. | | Deposit date: | 2012-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Domain Swapping of the Heme and N-Terminal alpha-Helix in Hydrogenobacter thermophilus Cytochrome c(552) Dimer
Biochemistry, 51, 2012
|
|
3WC8
 
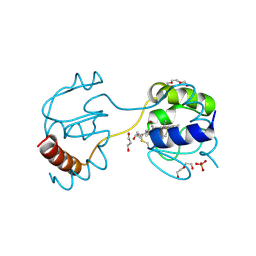 | | Dimeric horse cytochrome c obtained by refolding with desalting method | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Parui, P.P, Deshpande, M.S, Nagao, S, Kamikubo, H, Komori, H, Higuchi, Y, Kataoka, M, Hirota, S. | | Deposit date: | 2013-05-25 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Formation of Oligomeric Cytochrome c during Folding by Intermolecular Hydrophobic Interaction between N- and C-Terminal alpha-Helices
Biochemistry, 52, 2013
|
|
3WVR
 
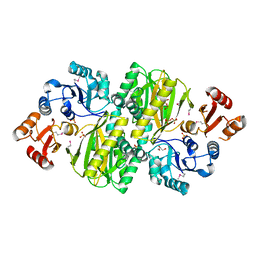 | | Structure of ATP grasp protein with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PGM1, ... | | Authors: | Matsui, T, Noike, M, Ooya, K, Sasaki, I, Hamano, Y, Maruyama, C, Ishikawa, J, Satoh, Y, Ito, H, Dairi, T, Morita, H. | | Deposit date: | 2014-06-04 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin.
Nat.Chem.Biol., 11, 2015
|
|
3WVQ
 
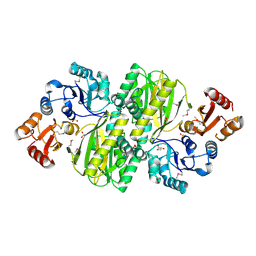 | | Structure of ATP grasp protein | | Descriptor: | GLYCEROL, PGM1, SULFATE ION | | Authors: | Matsui, T, Noike, M, Ooya, K, Sasaki, I, Hamano, Y, Maruyama, C, Ishikawa, J, Satoh, Y, Ito, H, Dairi, T, Morita, H. | | Deposit date: | 2014-06-04 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin.
Nat.Chem.Biol., 11, 2015
|
|
3WUI
 
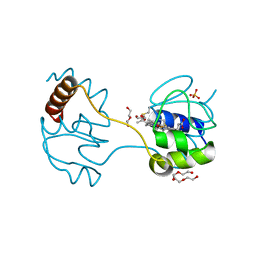 | | Dimeric horse cytochrome c formed by refolding from molten globule state | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Deshpande, M.S, Parui, P.P, Kamikubo, H, Yamanaka, M, Nagao, S, Komori, H, Kataoka, M, Higuchi, Y, Hirota, S. | | Deposit date: | 2014-04-25 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Formation of domain-swapped oligomer of cytochrome C from its molten globule state oligomer.
Biochemistry, 53, 2014
|
|
4ZID
 
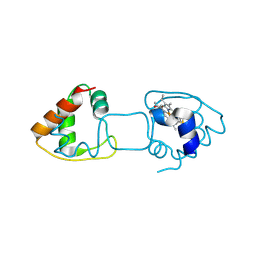 | | Dimeric Hydrogenobacter thermophilus cytochrome c552 obtained from Escherichia coli | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Hayashi, Y, Yamanaka, M, Nagao, S, Komori, H, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-04-28 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Domain swapping oligomerization of thermostable c-type cytochrome in E. coli cells
Sci Rep, 6, 2016
|
|
5AUL
 
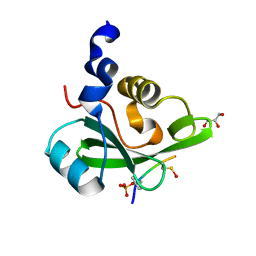 | | PI3K p85 C-terminal SH2 domain/CD28-derived peptide complex | | Descriptor: | GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, T-cell-specific surface glycoprotein CD28 | | Authors: | Inaba, S, Numoto, N, Morii, H, Ikura, T, Oda, M, Ito, N. | | Deposit date: | 2015-04-28 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structures and Thermodynamic Analysis Reveal Distinct Mechanisms of CD28 Phosphopeptide Binding to the Src Homology 2 (SH2) Domains of Three Adaptor Proteins
J. Biol. Chem., 292, 2017
|
|
5AUS
 
 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at C-terminal region | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
3SO2
 
 | | Chlorella dUTPase | | Descriptor: | Putative uncharacterized protein | | Authors: | Badalucco, L, Poudel, I, Natarajan, c, Yamanishi, M, Moriyama, H. | | Deposit date: | 2011-06-29 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6428 Å) | | Cite: | Crystallization of Chlorella deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3VKQ
 
 | | Assimilatory nitrite reductase (Nii3) - NO2 complex from tobbaco leaf analysed with middle X-ray dose | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
