4G10
 
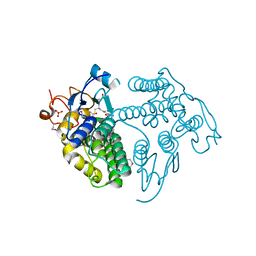 | | LigG from Sphingobium sp. SYK-6 is related to the glutathione transferase omega class | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase homolog, ... | | Authors: | Meux, E, Prosper, P, Masai, E, Mulliert Carlin, G, Dumarcay, S, Morel, M, Didierjean, C, Gelhaye, E, Favier, F. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Sphingobium sp. SYK-6 LigG involved in lignin degradation is structurally and biochemically related to the glutathione transferase omega class.
Febs Lett., 586, 2012
|
|
1ZTA
 
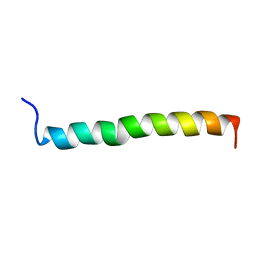 | | THE SOLUTION STRUCTURE OF A LEUCINE-ZIPPER MOTIF PEPTIDE | | Descriptor: | LEUCINE ZIPPER MONOMER | | Authors: | Saudek, V, Pastore, A, Castiglione Morelli, M.A, Frank, R, Gausepohl, H, Gibson, T. | | Deposit date: | 1990-10-11 | | Release date: | 1993-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a leucine-zipper motif peptide.
Protein Eng., 4, 1991
|
|
2BL6
 
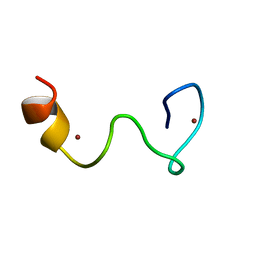 | | Solution structure of the Zn complex of EIAV NCp11(22-58) peptide, including two CCHC Zn-binding motifs. | | Descriptor: | NUCLEOCAPSID PROTEIN P11, ZINC ION | | Authors: | Amodeo, P, Castiglione-Morelli, M.A, Ostuni, A, Bavoso, A. | | Deposit date: | 2005-03-02 | | Release date: | 2006-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Features in Eiav Ncp11: A Lentivirus Nucleocapsid Protein with a Short Linker
Biochemistry, 45, 2006
|
|
1VIH
 
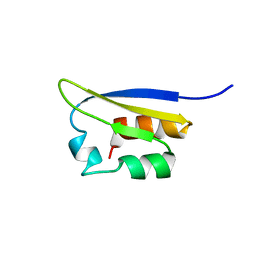 | | NMR STUDY OF VIGILIN, REPEAT 6, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
1VIG
 
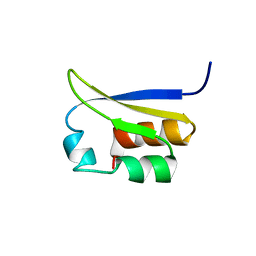 | | NMR STUDY OF VIGILIN, REPEAT 6, 40 STRUCTURES | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
1BMW
 
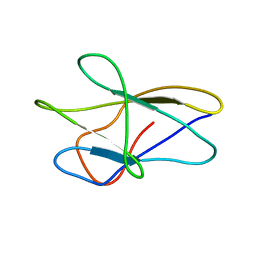 | | A fibronectin type III fold in plant allergens: The solution structure of Phl PII from timothy grass pollen, NMR, 38 STRUCTURES | | Descriptor: | POLLEN ALLERGEN PHL P2 | | Authors: | De Marino, S, Morelli, M.A.C, Fraternali, F, Tamborino, E, Vrtala, S, Dolecek, C, Arosio, P, Valenta, R, Pastore, A. | | Deposit date: | 1998-07-27 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An Immunoglobulin-Like Fold in a Major Plant Allergen: The Solution Structure of Phl P 2 from Timothy Grass Pollen.
Structure Fold.Des., 7, 1999
|
|
2GLH
 
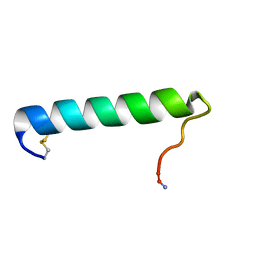 | | Solution Conformation of Salmon Calcitonin in Sodium Dodecyl Sulfate Micelles | | Descriptor: | Calcitonin-1 | | Authors: | Andreotti, G, Lopez-Mendez, B, Amodeo, P, Morelli, M.A, Nakamuta, H, Motta, A. | | Deposit date: | 2006-04-04 | | Release date: | 2006-06-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of salmon calcitonin bioactivity: the role of the Leu-based amphipathic alpha-helix.
J.Biol.Chem., 281, 2006
|
|
2GLG
 
 | | NMR structure of the [L23,A24]-sCT mutant | | Descriptor: | Calcitonin-1 | | Authors: | Andreotti, G, Lopez-Mendez, B, Amodeo, P, Morelli, M.A, Nakamuta, H, Motta, A. | | Deposit date: | 2006-04-04 | | Release date: | 2006-06-20 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of salmon calcitonin bioactivity: the role of the Leu-based amphipathic alpha-helix.
J.Biol.Chem., 281, 2006
|
|
2IWJ
 
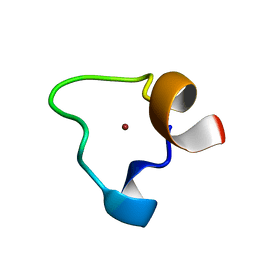 | | SOLUTION STRUCTURE OF THE ZN COMPLEX OF HIV-2 NCP(23-49) PEPTIDE, ENCOMPASSING PROTEIN CCHC-LINKER, DISTAL CCHC ZN-BINDING MOTIF AND C- TERMINAL TAIL. | | Descriptor: | GAG-POL POLYPROTEIN, ZINC ION | | Authors: | Amodeo, P, Castiglione Morelli, M.A, Ostuni, A, Cristinziano, P, Bavoso, A. | | Deposit date: | 2006-06-30 | | Release date: | 2007-10-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Features of the C-Terminal Zinc Finger Domain of the HIV-2 Nc Protein (Residues 23-49).
To be Published
|
|
