7V01
 
 | | Staphylococcus epidermidis RP62a CRISPR short effector complex with self RNA target and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZY
 
 | | Staphylococcus epidermidis RP62A CRISPR effector complex with non-self target RNA 2 | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZZ
 
 | | Staphylococcus epidermidis RP62a CRISPR tall effector complex | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7V02
 
 | | Staphylococcus epidermidis RP62A CRISPR short effector complex | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (4.97 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZW
 
 | | Staphylococcus epidermidis RP62a CRISPR effector subcomplex | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm4, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZX
 
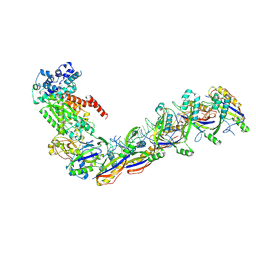 | | Staphylococcus epidermidis RP62a CRISPR effector subcomplex with non-self target RNA bound | | Descriptor: | CRISPR non-self RNA target, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7V00
 
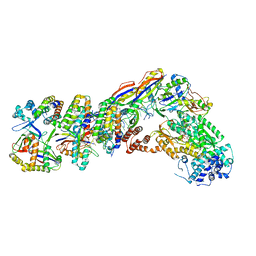 | | Staphylococcus epidermidis RP62a CRISPR tall effector complex with bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
3UD2
 
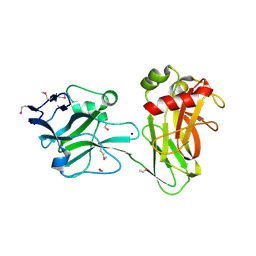 | | Crystal structure of Selenomethionine ZU5A-ZU5B protein domains of human erythrocyte ankyrin | | Descriptor: | Ankyrin-1, CHLORIDE ION, ETHANOL, ... | | Authors: | Yasunaga, M, Ipsaro, J.J, Mondragon, A. | | Deposit date: | 2011-10-27 | | Release date: | 2012-02-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structurally Similar but Functionally Diverse ZU5 Domains in Human Erythrocyte Ankyrin.
J.Mol.Biol., 417, 2012
|
|
4MTD
 
 | |
3UD1
 
 | |
2CSB
 
 | | Crystal structure of Topoisomerase V from Methanopyrus kandleri (61 kDa fragment) | | Descriptor: | MAGNESIUM ION, Topoisomerase V | | Authors: | Taneja, B, Patel, A, Slesarev, A, Mondragon, A. | | Deposit date: | 2005-05-21 | | Release date: | 2006-01-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases
Embo J., 25, 2006
|
|
2CSD
 
 | |
1CY2
 
 | |
1CY8
 
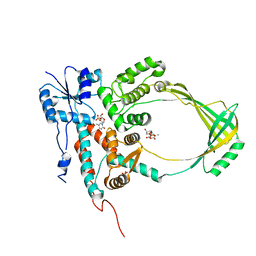 | | COMPLEX OF E.COLI DNA TOPOISOMERASE I WITH 5'-THYMIDINE MONOPHOSPHATE AND 3'-THYMIDINE MONOPHOSPHATE | | Descriptor: | DNA TOPOISOMERASE I, PHOSPHATE ION, THYMIDINE-3'-PHOSPHATE, ... | | Authors: | Feinberg, H, Changela, A, Mondragon, A. | | Deposit date: | 1999-08-31 | | Release date: | 2000-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Protein-nucleotide interactions in E. coli DNA topoisomerase I.
Nat.Struct.Biol., 6, 1999
|
|
1CY0
 
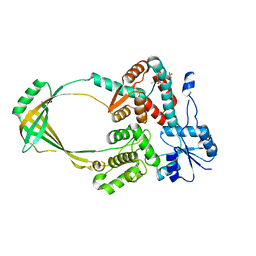 | |
6UGJ
 
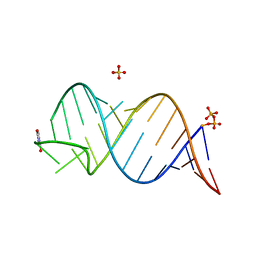 | |
6UGG
 
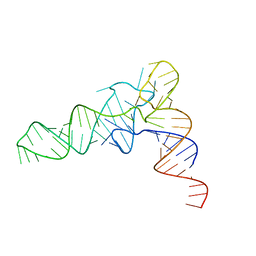 | | Structure of unmodified E. coli tRNA(Asp) | | Descriptor: | tRNAasp | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2019-09-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of an unmodified bacterial tRNA reveal intrinsic structural flexibility and plasticity as general properties of unbound tRNAs.
Rna, 26, 2020
|
|
6UGI
 
 | |
6VMY
 
 | | Structure of the B. subtilis cobalamin riboswitch | | Descriptor: | Adenosylcobalamin, B. subtilis cobalamin riboswitch, COBALT HEXAMMINE(III), ... | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of an atypical cobalamin riboswitch reveals RNA structural adaptability as basis for promiscuous ligand binding.
Nucleic Acids Res., 48, 2020
|
|
2F4Q
 
 | |
3F57
 
 | |
3F59
 
 | |
1CYY
 
 | |
6N1R
 
 | | Tetrahedral oligomeric complex of GyrA N-terminal fragment, solved by cryoEM in tetrahedral symmetry | | Descriptor: | DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
6CWX
 
 | | Crystal structure of human ribonuclease P/MRP proteins Rpp20/Rpp25 | | Descriptor: | FORMIC ACID, Ribonuclease P protein subunit p20, Ribonuclease P protein subunit p25, ... | | Authors: | Chan, C.W, Kiesel, B.R, Mondragon, A. | | Deposit date: | 2018-03-31 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Human Rpp20/Rpp25 Reveals Quaternary Level Adaptation of the Alba Scaffold as Structural Basis for Single-stranded RNA Binding.
J. Mol. Biol., 430, 2018
|
|
