8A39
 
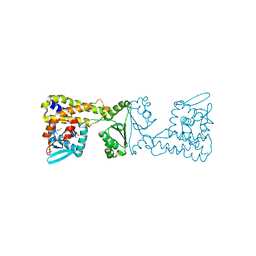 | | Crystal Structure of PaaX from Escherichia coli W | | Descriptor: | DNA-binding transcriptional repressor of phenylacetic acid degradation, aryl-CoA responsive, GLYCEROL, ... | | Authors: | Molina, R, Alba-Perez, A, Hermoso, J.A. | | Deposit date: | 2022-06-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of PaaX, the main repressor of the phenylacetate degradation pathway in Escherichia coli W: A novel fold of transcription regulator proteins.
Int.J.Biol.Macromol., 254, 2024
|
|
4UT0
 
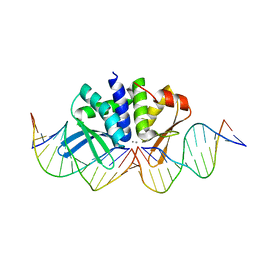 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 10 DAYS INCUBATION IN 5MM MN (STATE 7) | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*CP)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*CP)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-07-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
9H68
 
 | |
7Z55
 
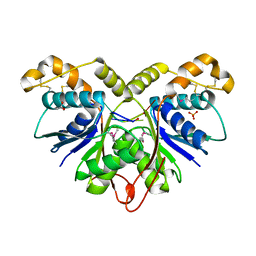 | | Crystal Structure of the Ring Nuclease 0455 from Sulfolobus islandicus (Sis0455) in complex with its substrate | | Descriptor: | ACETATE ION, CRISPR-associated protein, Cyclic RNA cA4, ... | | Authors: | Molina, R, Martin-Garcia, R, Lopez-Mendez, B, Jensen, A.L.G, Marchena-Hurtado, J, Stella, S, Montoya, G. | | Deposit date: | 2022-03-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | Molecular basis of cyclic tetra-oligoadenylate processing by small standalone CRISPR-Cas ring nucleases.
Nucleic Acids Res., 50, 2022
|
|
7Z56
 
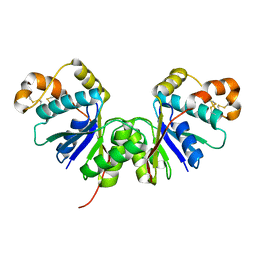 | | Crystal Structure of the Ring Nuclease 0455 from Sulfolobus islandicus (Sis0455) in its apo form | | Descriptor: | CRISPR-associated protein | | Authors: | Molina, R, Martin-Garcia, R, Lopez-Mendez, B, Jensen, A.L.G, Marchena-Hurtado, J, Stella, S, Montoya, G. | | Deposit date: | 2022-03-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular basis of cyclic tetra-oligoadenylate processing by small standalone CRISPR-Cas ring nucleases.
Nucleic Acids Res., 50, 2022
|
|
6FB0
 
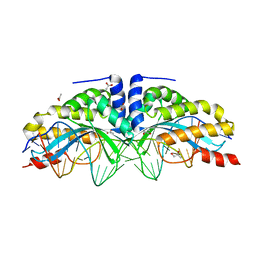 | |
6FB5
 
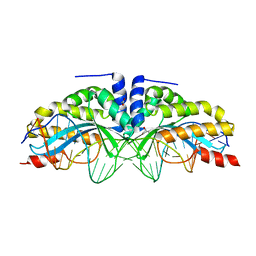 | |
6FB1
 
 | |
6FB8
 
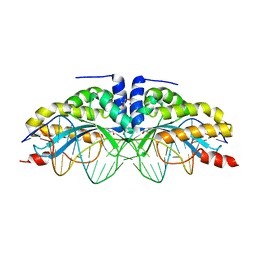 | |
6FB2
 
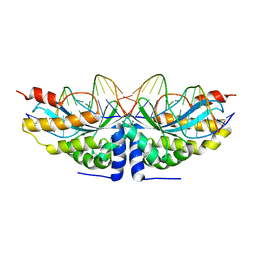 | |
6FB7
 
 | |
6R9R
 
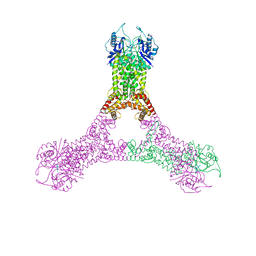 | | Crystal structure of Csx1 in complex with cyclic oligoadenylate cOA4 conformation 2 | | Descriptor: | CRISPR-associated (Cas) DxTHG family, circular RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Molina, R, Montoya, G, Sofos, N, Stella, S. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Csx1-cOA4complex reveals the basis of RNA decay in Type III-B CRISPR-Cas.
Nat Commun, 10, 2019
|
|
6R7B
 
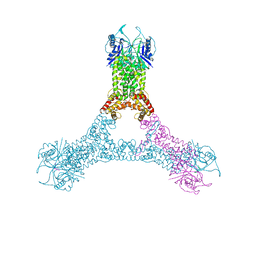 | | Crystal structure of Csx1 in complex with cyclic oligoadenylate cOA4 conformation 1 | | Descriptor: | CRISPR-associated (Cas) DxTHG family, RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Molina, R, Montoya, G, Sofos, N, Stella, S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure of Csx1-cOA4complex reveals the basis of RNA decay in Type III-B CRISPR-Cas.
Nat Commun, 10, 2019
|
|
4UN8
 
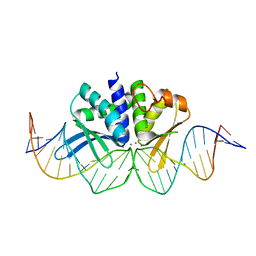 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 1H INCUBATION IN 5MM MN (STATE 2) | | Descriptor: | 25MER, HOMING ENDONUCLEASE I-DMOI, MANGANESE (II) ION | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UN9
 
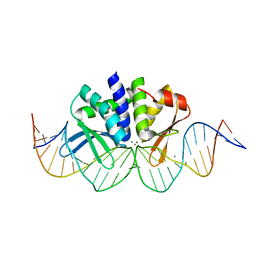 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 8H INCUBATION IN 5MM MN (STATE 3) | | Descriptor: | 25MER, HOMING ENDONUCLEASE I-DMOI, MANGANESE (II) ION | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UNA
 
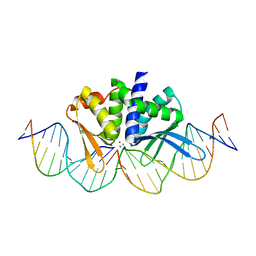 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 2 DAYS INCUBATION IN 5MM MN (STATE 4) | | Descriptor: | 25MER, 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UNC
 
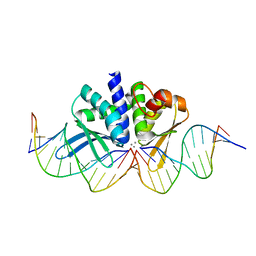 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 8 DAYS INCUBATION IN 5MM MN (STATE 6) | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*A)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UNB
 
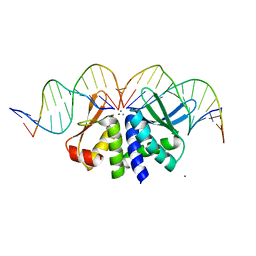 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 6 DAYS INCUBATION IN 5MM MN (STATE 5) | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*A)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UN7
 
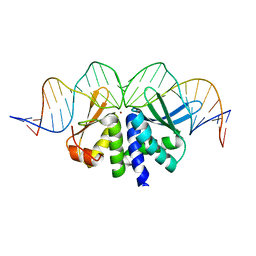 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA BEFORE INCUBATION IN 5MM MN (STATE 1) | | Descriptor: | 25MER, HOMING ENDONUCLEASE I-DMOI, ZINC ION | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6QZQ
 
 | |
6QZT
 
 | |
7PQ3
 
 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in complex with its post-catalytic reaction product | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQ2
 
 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in its apo form | | Descriptor: | CRISPR-associated protein, APE2256 family, CRISPR Ring Nuclease | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQA
 
 | | Crystal Structure of the Ring Nuclease 0811 mutant-S12G/K169G from Sulfolobus islandicus (Sis0811) | | Descriptor: | CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQ6
 
 | | Crystal Structure of the Ring Nuclease 0811 mutant-S12A from Sulfolobus islandicus (Sis0811) | | Descriptor: | CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
