2C2P
 
 | | The crystal structure of mismatch specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans | | Descriptor: | ACETATE ION, G/U MISMATCH-SPECIFIC DNA GLYCOSYLASE | | Authors: | Moe, E, Leiros, I, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of Mismatch Specific Uracil-DNA Glycosylase (Mug) from Deinococcus Radiodurans Reveals a Novel Catalytic Residue and Broad Substrate Specificity
J.Biol.Chem., 281, 2006
|
|
2C2Q
 
 | | The crystal structure of mismatch specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans. Inactive mutant Asp93Ala. | | Descriptor: | ACETATE ION, G/U MISMATCH-SPECIFIC DNA GLYCOSYLASE | | Authors: | Moe, E, Leiros, I, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Mismatch Specific Uracil-DNA Glycosylase (Mug) from Deinococcus Radiodurans Reveals a Novel Catalytic Residue and Broad Substrate Specificity
J.Biol.Chem., 281, 2006
|
|
2YG9
 
 | | Structure of an unusual 3-Methyladenine DNA Glycosylase II (Alka) from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, DNA-3-methyladenine glycosidase II, putative, ... | | Authors: | Moe, E, Hall, D.R, Leiros, I, Talstad, V, Timmins, J, McSweeney, S. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
1YUO
 
 | | Optimisation of the surface electrostatics as a strategy for cold adaptation of uracil-DNA N-glycosylase (UNG)from atlantic cod (Gadus morhua) | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Moe, E, Leiros, I, Riise, E.K, Olufsen, M, Lanes, O, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimisation of the surface electrostatics as a strategy for cold adaptation of uracil-DNA N-glycosylase (UNG) from Atlantic cod (Gadus morhua)
J.Mol.Biol., 343, 2004
|
|
2YG8
 
 | | Structure of an unusual 3-Methyladenine DNA Glycosylase II (Alka) from Deinococcus radiodurans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA-3-methyladenine glycosidase II, ... | | Authors: | Moe, E, Hall, D.R, Leiros, I, Talstad, V, Timmins, J, McSweeney, S. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
6GEH
 
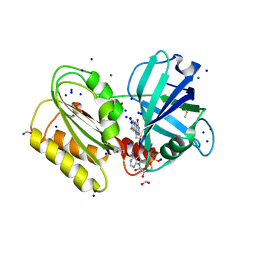 | | Structure and reactivity of a siderophore-interacting protein from the marine bacterium Shewanella reveals unanticipated functional versatility. | | Descriptor: | DIMETHYL SULFOXIDE, FAD-binding 9, siderophore-interacting domain protein, ... | | Authors: | Trindade, I.B, Silva, J.P.M, Matias, P, Moe, E. | | Deposit date: | 2018-04-26 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and reactivity of a siderophore-interacting protein from the marine bacteriumShewanellareveals unanticipated functional versatility.
J. Biol. Chem., 294, 2019
|
|
2BOO
 
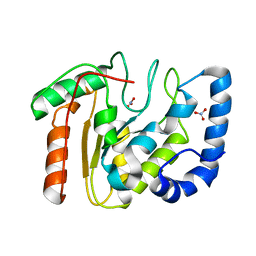 | | The crystal structure of Uracil-DNA N-Glycosylase (UNG) from Deinococcus radiodurans. | | Descriptor: | NITRATE ION, URACIL-DNA GLYCOSYLASE | | Authors: | Leiros, I, Moe, E, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-04-13 | | Release date: | 2005-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Uracil-DNA N-Glycosylase (Ung) from Deinococcus Radiodurans.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8A5G
 
 | |
8A5F
 
 | |
8A5C
 
 | |
7QP5
 
 | | Crystal Structure of E. coli FhuF | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferric iron reductase protein FhuF | | Authors: | Trindade, I.B, Rollo, F, Matias, P.M, Moe, E, Louro, R.O. | | Deposit date: | 2022-01-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of a novel ferredoxin: FhuF, a ferric-siderophore reductase from E. coli K-12 with a novel 2Fe-2S cluster coordination
Biorxiv, 2023
|
|
2CE4
 
 | | Manganese Superoxide Dismutase (Mn-SOD) from Deinococcus radiodurans | | Descriptor: | MANGANESE (II) ION, SUPEROXIDE DISMUTASE [MN] | | Authors: | Dennis, R, Micossi, E, McCarthy, J, Moe, E, Gordon, E, Leonard, G, McSweeney, S. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the manganese superoxide dismutase from Deinococcus radiodurans in two crystal forms.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 62, 2006
|
|
2CDY
 
 | | Manganese Superoxide Dismutase (Mn-SOD) from Deinococcus radiodurans | | Descriptor: | MANGANESE (II) ION, SUPEROXIDE DISMUTASE [MN] | | Authors: | Dennis, R, Micossi, E, McCarthy, J, Moe, E, Gordon, E, Leonard, G, McSweeney, S. | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the manganese superoxide dismutase from Deinococcus radiodurans in two crystal forms.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 62, 2006
|
|
4LYL
 
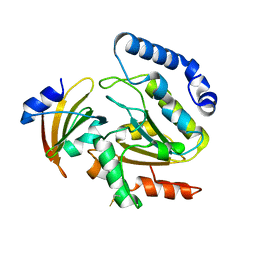 | | Crystal structure of uracil-DNA glycosylase from cod (Gadus morhua) in complex with the proteinaceous inhibitor UGI | | Descriptor: | Uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Assefa, N.G, Niiranen, L.M.K, Johnson, K.A, Leiros, H.-K.S, Smalas, A.O, Willassen, N.P, Moe, E. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and biophysical analysis of interactions between cod and human uracil-DNA N-glycosylase (UNG) and UNG inhibitor (Ugi).
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TRT
 
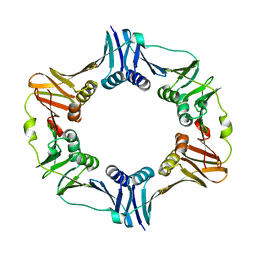 | |
4UOB
 
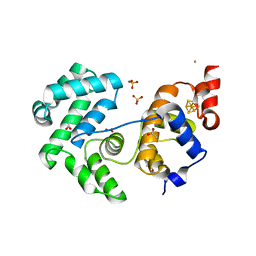 | | Crystal structure of Deinococcus radiodurans Endonuclease III-3 | | Descriptor: | ACETATE ION, COBALT (II) ION, ENDONUCLEASE III-3, ... | | Authors: | Sarre, A, Okvist, M, Klar, T, Hall, D, Smalas, A.O, McSweeney, S, Timmins, J, Moe, E. | | Deposit date: | 2014-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural and Functional Characterization of Two Unusual Endonuclease III Enzymes from Deinococcus Radiodurans.
J.Struct.Biol., 191, 2015
|
|
4UNF
 
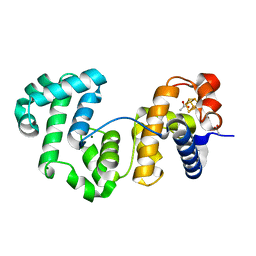 | | Crystal structure of Deinococcus radiodurans Endonuclease III-1 | | Descriptor: | ACETATE ION, ENDONUCLEASE III-1, IRON/SULFUR CLUSTER, ... | | Authors: | Sarre, A, Okvist, M, Klar, T, Hall, D, Smalas, A.O, McSweeney, S, Timmins, J, Moe, E. | | Deposit date: | 2014-05-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of Two Unusual Endonuclease III Enzymes from Deinococcus Radiodurans.
J.Struct.Biol., 191, 2015
|
|
4UQM
 
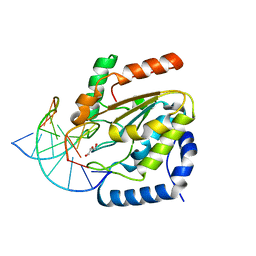 | | Crystal structure determination of uracil-DNA N-glycosylase (UNG) from Deinococcus radiodurans in complex with DNA - new insights into the role of the Leucine-loop for damage recognition and repair | | Descriptor: | 5'-D(*CP*CP*TP*AP*TP*CP*CP*AP*AAB*GP*TP*CP*TP*CP*CP*G)-3', 5'-D(*GP*CP*GP*GP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*AP*G)-3', CHLORIDE ION, ... | | Authors: | Pedersen, H.L, Johnson, K.A, McVey, C.E, Leiros, I, Moe, E. | | Deposit date: | 2014-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure determination of uracil-DNA N-glycosylase from Deinococcus radiodurans in complex with DNA.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4V14
 
 | |
1OKB
 
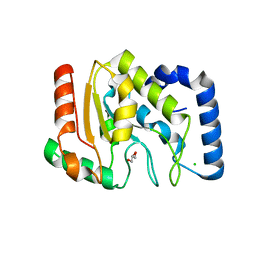 | | crystal structure of Uracil-DNA glycosylase from Atlantic cod (Gadus morhua) | | Descriptor: | CHLORIDE ION, GLYCEROL, URACIL-DNA GLYCOSYLASE | | Authors: | Leiros, I, Moe, E, Lanes, O, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2003-07-21 | | Release date: | 2004-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Uracil-DNA Glycosylase from Atlantic Cod (Gadus Morhua) Reveals Cold-Adaptation Features
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2PU3
 
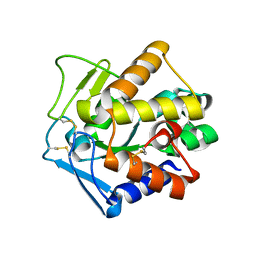 | | Structural adaptation of endonuclease I from the cold-adapted and halophilic bacterium Vibrio salmonicida | | Descriptor: | CHLORIDE ION, Endonuclease I, MAGNESIUM ION | | Authors: | Altermark, B, Helland, R, Moe, E, Willassen, N.P, Smalas, A.O. | | Deposit date: | 2007-05-08 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural adaptation of endonuclease I from the cold-adapted and halophilic bacterium Vibrio salmonicida.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
8AK4
 
 | | Structure of the C-terminally truncated NAD+-dependent DNA ligase from the poly-extremophile Deinococcus radiodurans | | Descriptor: | DNA ligase, MANGANESE (II) ION, ZINC ION | | Authors: | Fernandes, A, Williamson, A.K, Matias, P.M, Moe, E. | | Deposit date: | 2022-07-29 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structure/function studies of the NAD + -dependent DNA ligase from the poly-extremophile Deinococcus radiodurans reveal importance of the BRCT domain for DNA binding.
Extremophiles, 27, 2023
|
|
8C4L
 
 | |
3TKB
 
 | | crystal structure of human uracil-DNA glycosylase D183G/K302R mutant | | Descriptor: | IMIDAZOLE, Uracil-DNA glycosylase | | Authors: | Assefa, N.G, Niiranen, L, Willassen, N.P, Smalas, A.O, Moe, E. | | Deposit date: | 2011-08-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thermal unfolding studies of cold adapted uracil-DNA N-glycosylase (UNG) from Atlantic cod (Gadus morhua). A comparative study with human UNG.
Comp.Biochem.Physiol. B: Biochem.Mol.Biol., 161, 2012
|
|
2V28
 
 | | Apo structure of the cold active phenylalanine hydroxylase from Colwellia psychrerythraea 34H | | Descriptor: | PHENYLALANINE-4-HYDROXYLASE, SULFATE ION | | Authors: | Leiros, H.-K.S, Pey, A.L, Innselset, M, Moe, E, Leiros, I, Steen, I.H, Martinez, A. | | Deposit date: | 2007-06-04 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Phenylalanine Hydroxylase from Colwellia Psychrerythraea 34H, a Monomeric Cold Active Enzyme with Local Flexibility Around the Active Site and High Overall Stability.
J.Biol.Chem., 282, 2007
|
|
