8IUB
 
 | |
8IUC
 
 | |
3A64
 
 | | Crystal structure of CcCel6C, a glycoside hydrolase family 6 enzyme, from Coprinopsis cinerea | | Descriptor: | Cellobiohydrolase, MAGNESIUM ION | | Authors: | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
8R84
 
 | | pentameric IgMFc-AIM complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CD5 antigen-like, ... | | Authors: | Chen, Q, Arai, S, Miyazaki, T, Rosenthal, P. | | Deposit date: | 2023-11-28 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM reveals structural basis for human AIM/CD5L recognition of polymeric immunoglobulin M
Nat Commun, 15, 2024
|
|
8R83
 
 | | pentameric IgMFc-AIM complex global refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, Q, Arai, S, Miyazaki, T, Rosenthal, P. | | Deposit date: | 2023-11-28 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM reveals structural basis for human AIM/CD5L recognition of polymeric immunoglobulin M
Nat Commun, 15, 2024
|
|
3WPZ
 
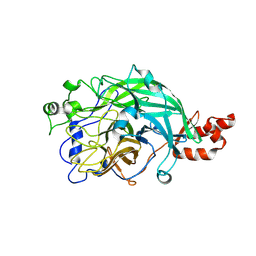 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/S200T/F447P/F470Y/P500S | | Descriptor: | Beta-fructofuranosidase | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WJJ
 
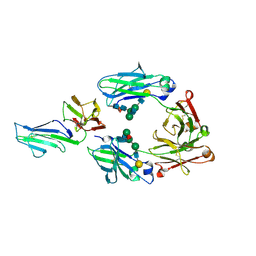 | | Crystal structure of IIb selective Fc variant, Fc(P238D), in complex with FcgRIIb | | Descriptor: | Ig gamma-1 chain C region, Low affinity immunoglobulin gamma Fc region receptor II-b, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kadono, S, Mimoto, F, Katada, H, Igawa, T, Kuramochi, T, Muraoka, M, Wada, Y, Haraya, K, Miyazaki, T, Hattori, K. | | Deposit date: | 2013-10-10 | | Release date: | 2013-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineered antibody Fc variant with selectively enhanced Fc gamma RIIb binding over both Fc gamma RIIaR131 and Fc gamma RIIaH131.
Protein Eng.Des.Sel., 26, 2013
|
|
3A9B
 
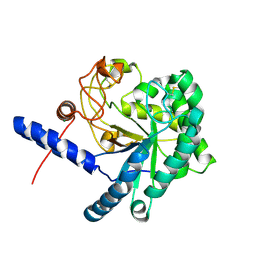 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with cellobiose | | Descriptor: | Cellobiohydrolase, MAGNESIUM ION, beta-D-glucopyranose, ... | | Authors: | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3ABX
 
 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with p-nitrophenyl beta-D-cellotrioside | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, Cellobiohydrolase, MAGNESIUM ION | | Authors: | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-12-24 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
8J53
 
 | |
8J50
 
 | |
8J52
 
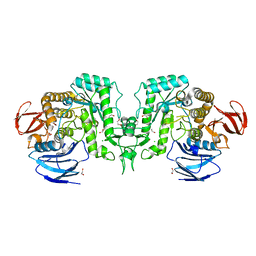 | | Crystal structure of Flavihumibacter petaseus GH31 alpha-galactosidase mutant D304A in complex with alpha-1,4-galactobiose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GH31 alpha-galactosidase, ... | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2023-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analysis of bacterial GH31 alpha-galactosidases specific for alpha-(1→4)-galactobiose.
Febs J., 290, 2023
|
|
8J51
 
 | |
3WZ1
 
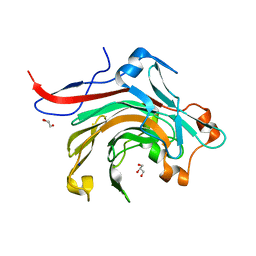 | | Catalytic domain of beta-agarase from Microbulbifer thermotolerans JAMB-A94 | | Descriptor: | Agarase, GLYCEROL, SODIUM ION | | Authors: | Takagi, E, Hatada, Y, Akita, M, Ohta, Y, Yokoi, G, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the catalytic domain of a GH16 beta-agarase from a deep-sea bacterium, Microbulbifer thermotolerans JAMB-A94
Biosci.Biotechnol.Biochem., 79, 2015
|
|
3APF
 
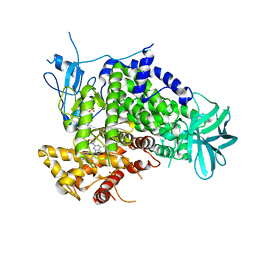 | | Crystal structure of human PI3K-gamma in complex with CH5039699 | | Descriptor: | 3-[7-(1H-benzimidazol-5-yl)-2-(morpholin-4-yl)-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl]phenol, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Nakamura, M, Fukami, T.A, Miyazaki, T, Yoshida, M. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery and biological activity of a novel class I PI3K inhibitor, CH5132799
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3APC
 
 | | Crystal structure of human PI3K-gamma in complex with CH5132799 | | Descriptor: | 5-(7-Methanesulfonyl-2-morpholin-4-yl-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl)-pyrimidin-2-ylamine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Nakamura, M, Fukami, T.A, Miyazaki, T, Yoshida, M. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.544 Å) | | Cite: | Discovery and biological activity of a novel class I PI3K inhibitor, CH5132799
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3APD
 
 | | Crystal structure of human PI3K-gamma in complex with CH5108134 | | Descriptor: | 5-(2-Morpholin-4-yl-7-pyridin-3-yl-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl)-pyrimidin-2-ylamine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Nakamura, M, Fukami, T.A, Miyazaki, T, Yoshida, M. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and biological activity of a novel class I PI3K inhibitor, CH5132799
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7WLG
 
 | | Cryo-EM structure of GH31 alpha-1,3-glucosidase from Lactococcus lactis subsp. cremoris | | Descriptor: | Alpha-xylosidase | | Authors: | Ikegaya, M, Moriya, T, Adachi, N, Kawasaki, M, Park, E.Y, Miyazaki, T. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJE
 
 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerotetraose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJF
 
 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with kojibiose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-2)-alpha-D-glucopyranose | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJD
 
 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerotriose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, ... | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJ9
 
 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase, P21 space group | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, Xylitol | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJB
 
 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose, ... | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJC
 
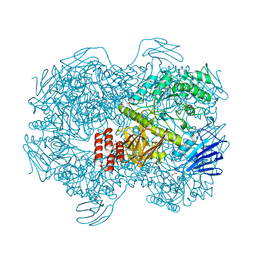 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJA
 
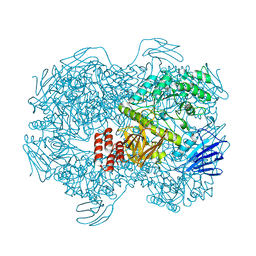 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase, P6322 space group | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
