2A35
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA4017 from Pseudomonas aeruginosa PAO1, Possible Epimerase | | Descriptor: | hypothetical protein PA4017 | | Authors: | Zhang, R, Xu, L, Cuff, M, Savchenko, A, Cymborowski, M, Minor, W, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-23 | | Release date: | 2005-08-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA4017 from
Pseudomonas aeruginosa PAO1
To be Published
|
|
2AZ4
 
 | | Crystal Structure of a Protein of Unknown Function from Enterococcus faecalis V583 | | Descriptor: | ZINC ION, hypothetical protein EF2904 | | Authors: | Zhang, R, Maltseva, N, Moy, S, Collart, F, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-09 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of a hypothetical protein from Enterococcus faecalis V583
To be Published
|
|
2B2T
 
 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and phosphothreonine 3 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 tail | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
4X9O
 
 | | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae soaked with octanoyl-CoA: conformational changes without clearly bound substrate | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2 | | Authors: | Hou, J, Cooper, D.R, Shabalin, I.G, Grabowski, M, Shumilin, I, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
to be published
|
|
4XCV
 
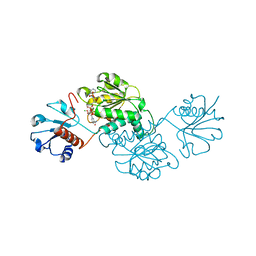 | | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, NADP-dependent 2-hydroxyacid dehydrogenase, ... | | Authors: | Langner, K.M, Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Stead, M, Hillerich, B.S, Chowdhury, S, Hammonds, J, Zimmerman, M.D, Al Obadi, N, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-18 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH
to be published
|
|
4XK4
 
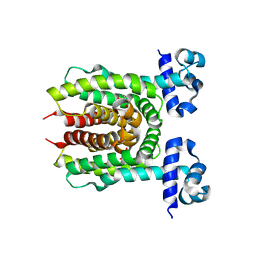 | | E. coli transcriptional regulator RUTR with dihydrouracil | | Descriptor: | DIHYDROPYRIMIDINE-2,4(1H,3H)-DIONE, HTH-type transcriptional regulator RutR | | Authors: | Shumilin, I.A, Cooper, D.R, Shabalin, I.G, Grabowski, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-01-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | E. COLI TRANSCRIPTIONAL REGULATOR RUTR WITH DIHYDROURACIL
to be published
|
|
4XA8
 
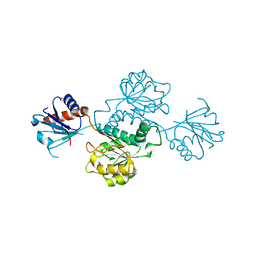 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding | | Authors: | Handing, K.B, Gasiorowska, O.A, Shabalin, I.G, Cymborowski, M.T, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-12 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2.
to be published
|
|
4X9K
 
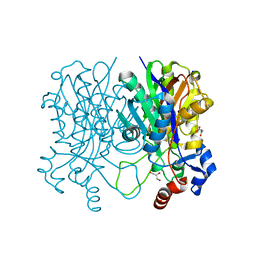 | | Beta-ketoacyl-acyl carrier protein synthase III-2 (FabH2)(C113A) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, GLYCEROL, MALONATE ION, ... | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Chordia, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
to be published
|
|
4EUY
 
 | | Crystal structure of thioredoxin-like protein BCE_0499 from Bacillus cereus ATCC 10987 | | Descriptor: | Uncharacterized protein | | Authors: | Shabalin, I.G, Kagan, O, Chruszcz, M, Grabowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of thioredoxin-like protein BCE_0499 from
Bacillus cereus
To be Published
|
|
4XOX
 
 | | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae | | Descriptor: | 3-oxoacyl-ACP synthase | | Authors: | Hou, J, Grabowski, M, Cymborowski, M, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae
to be published
|
|
1WQ6
 
 | | The tetramer structure of the nervy homolgy two (NHR2) domain of AML1-ETO is critical for AML1-ETO'S activity | | Descriptor: | AML1-ETO | | Authors: | Liu, Y, Cheney, M.D, Chruszcz, M, Lukasik, S.M, Hartman, K.L, Laue, T.M, Dauter, Z, Minor, W, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2004-09-23 | | Release date: | 2005-10-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity
Cancer Cell, 9, 2006
|
|
4YEO
 
 | | Triclinic HEWL co-crystallised with cisplatin, studied at a data collection temperature of 150K - new refinement | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cisplatin, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5VDB
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with bisubstrate analog 3 | | Descriptor: | (3R,5S,9R,26S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14,20-trioxo-26-({[(phenylacetyl)amino]acetyl}amino)-2,4,6-trioxa-18-thia-11,15,21-triaza-3,5-diphosphaheptacosan-27-oic acid 3,5-dioxide (non-preferred name), SULFATE ION, acetyltransferase PA4794 | | Authors: | Majorek, K.A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-04-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
FEBS Lett., 591, 2017
|
|
4ZNZ
 
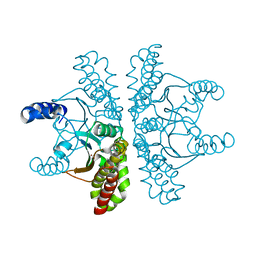 | | Crystal structure of Escherichia coli carbonic anhydrase (YadF) in complex with Zn - artifact of purification | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Gasiorowska, O.A, Niedzialkowska, E, Porebski, P.J, Handing, K.B, Shabalin, I.G, Cymborowski, M.T, Minor, W. | | Deposit date: | 2015-05-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
4YDX
 
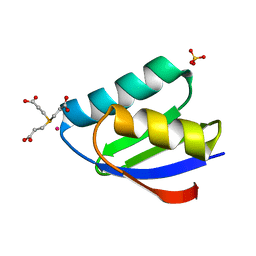 | | Crystal structure of cisplatin bound to a human copper chaperone (monomer) - new refinement | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Copper transport protein ATOX1, PLATINUM (II) ION, ... | | Authors: | Shabalin, I.G, Boal, A.K, Dauter, Z, Jaskolski, M, Minor, W, Rosenzweig, A.C, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEA
 
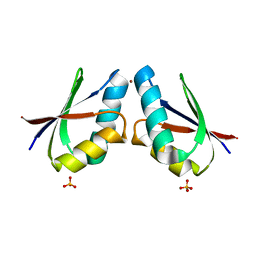 | | Crystal structure of cisplatin bound to a human copper chaperone (dimer) - new refinement | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, SULFATE ION | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEM
 
 | | Carboplatin binding to HEWL in NaBr crystallisation conditions studied at an X-ray wavelength of 0.9163A - new refinement | | Descriptor: | ACETATE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZQB
 
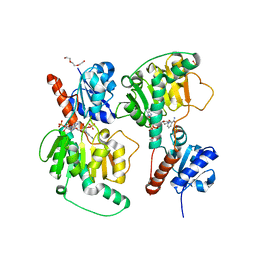 | | Crystal structure of NADP-dependent dehydrogenase from Rhodobactersphaeroides in complex with NADP and sulfate | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kowiel, M, Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Porebski, P.J, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-08 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of NADP-dependent dehydrogenase from Rhodobactersphaeroides in complex with NADP and sulfate
to be published
|
|
4YYC
 
 | | Crystal structure of trimethylamine methyltransferase from Sinorhizobium meliloti in complex with unknown ligand | | Descriptor: | CHLORIDE ION, Putative trimethylamine methyltransferase, UNKNOWN LIGAND | | Authors: | Shabalin, I.G, Porebski, P.J, Gasiorowska, O.A, Handing, K.B, Niedzialkowska, E, Cymborowski, M.T, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
4XAP
 
 | | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 | | Descriptor: | Aldo-keto reductase | | Authors: | Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021
to be published
|
|
4GQA
 
 | | Crystal structure of NAD binding oxidoreductase from Klebsiella pneumoniae | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NAD binding Oxidoreductase, ... | | Authors: | Osinski, S, Majorek, K.A, Niedzialkowska, E, Osinski, T, Porebski, P.J, Nawar, A, Hammonds, J, Hillerich, B, Seidel, R, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-22 | | Release date: | 2012-09-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of NAD binding oxidoreductase from Klebsiella pneumoniae (CASP Target)
To be Published
|
|
4HE5
 
 | | Crystal structure of the selenomethionine variant of the C-terminal domain of Geobacillus thermoleovorans putative U32 peptidase | | Descriptor: | Peptidase family U32, SULFATE ION | | Authors: | Trillo-Muyo, S, Jasilionis, A, Domagalski, M.J, Chruszcz, M, Minor, W, Kuisiene, N, Arolas, J.L, Sola, M, Gomis-Ruth, F.X. | | Deposit date: | 2012-10-03 | | Release date: | 2012-11-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ultratight crystal packing of a 10 kDa protein.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4Z0P
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in complex with NADPH and oxalate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sroka, P, Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Porebski, P.J, Hillerich, B.S, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
1Y89
 
 | | Crystal Structure of devB protein | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DI(HYDROXYETHYL)ETHER, NONAETHYLENE GLYCOL, ... | | Authors: | Lazarski, K, Cymborowski, M, Chruszcz, M, Zheng, H, Zhang, R, Lezondra, L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of devB protein
To be Published
|
|
4HE6
 
 | | Crystal structure of the C-terminal domain of Geobacillus thermoleovorans putative U32 peptidase | | Descriptor: | ACETATE ION, Peptidase family U32, UNKNOWN ATOM OR ION | | Authors: | Trillo-Muyo, S, Jasilionis, A, Domagalski, M.J, Chruszcz, M, Minor, W, Kuisiene, N, Arolas, J.L, Sola, M, Gomis-Ruth, F.X. | | Deposit date: | 2012-10-03 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ultratight crystal packing of a 10 kDa protein.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
