3CM1
 
 | |
2FQ4
 
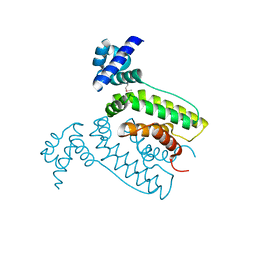 | | The crystal structure of the transcriptional regulator (TetR family) from Bacillus cereus | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Zhang, R, Wu, R, Moy, S, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-17 | | Release date: | 2006-02-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The crystal structure of the transcriptional regulator (TetR family) from Bacillus cereus
To be Published
|
|
3KZP
 
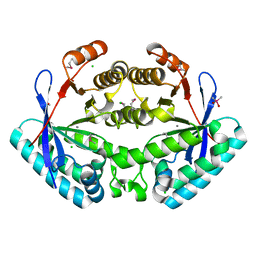 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Listaria monocytigenes | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Zimmerman, M.D, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Listaria monocytigenes
To be Published
|
|
3UED
 
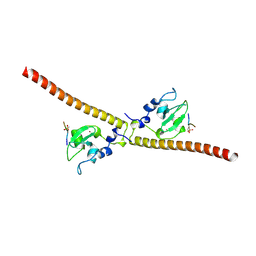 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3 (C2 space group). | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
7KPP
 
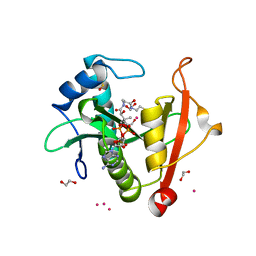 | | Structure of the E102A mutant of a GNAT superfamily PA3944 acetyltransferase | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, COENZYME A, ... | | Authors: | Czub, M.P, Porebski, P.J, Majorek, K.A, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
7KPS
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with AcCoA | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL COENZYME *A, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Reidl, C.T, Becker, D.P, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
7KYE
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with CHES | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetyltransferase PA3944, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with CHES
To Be Published
|
|
3UEE
 
 | | Crystal structure of human Survivin K62A mutant bound to N-terminal histone H3 | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3KKD
 
 | | Structure of a putative tetr transcriptional regulator (pa3699) from pseudomonas aeruginosa pa01 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Filippova, E.V, Chruszcz, M, Cymborowski, M, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-05 | | Release date: | 2009-12-15 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Putative TetR Transcriptional Regulator (PA3699) from Pseudomonas Aeruginosa PA01
To be Published
|
|
1HVY
 
 | | Human thymidylate synthase complexed with dUMP and Raltitrexed, an antifolate drug, is in the closed conformation | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, BETA-MERCAPTOETHANOL, THYMIDYLATE SYNTHASE, ... | | Authors: | Phan, J, Koli, S, Minor, W, Dunlap, R.B, Berger, S.H, Lebioda, L. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human thymidylate synthase is in the closed conformation when complexed with dUMP and raltitrexed, an antifolate drug
Biochemistry, 40, 2001
|
|
3N73
 
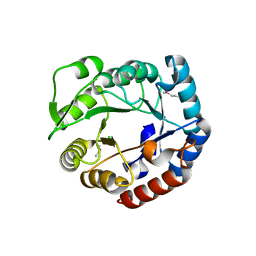 | | Crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus cereus | | Descriptor: | CHLORIDE ION, Putative 4-hydroxy-2-oxoglutarate aldolase | | Authors: | Cabello, R, Chruszcz, M, Xu, X, Zimmerman, M.D, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus cereus
To be Published
|
|
6CIA
 
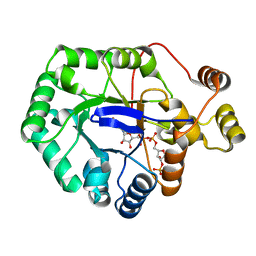 | | Crystal structure of aldo-keto reductase from Klebsiella pneumoniae in complex with NADPH. | | Descriptor: | Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lipowska, J, Leung, E.S, Shabalin, I.G, Grabowski, M, Almo, S.C, Satchell, K.J, Joachimiak, A, Lewinski, K, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of of aldo-keto reductase from Klebsiella pneumoniae in complex with NADPH.
to be published
|
|
1YPV
 
 | |
2M3W
 
 | |
5W8W
 
 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 - new refinement | | Descriptor: | Metallo-beta-lactamase type 2, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Gonzalez, J.M, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Wlodawer, A, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2017-06-22 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
7XA9
 
 | | Structure of Arabidopsis thaliana CLCa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Ji, S, Jin, H, Kaiming, Z, Mingxing, W, Shanshan, L, Long, C. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the plant nitrate transporter AtCLCa reveals characteristics of the anion-binding site and the ATP-binding pocket.
J.Biol.Chem., 299, 2023
|
|
4FCG
 
 | | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Zimmerman, M.D, Minor, W, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-13 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL)
To be Published
|
|
2Q24
 
 | | Crystal structure of TetR transcriptional regulator SCO0520 from Streptomyces coelicolor | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative tetR family transcriptional regulator | | Authors: | Cymborowski, M, Chruszcz, M, Koclega, K.D, Filippova, E.V, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-25 | | Release date: | 2007-07-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative transcriptional regulator SCO0520 from Streptomyces coelicolor A3(2) reveals an unusual dimer among TetR family proteins.
J.Struct.Funct.Genom., 12, 2011
|
|
3KXR
 
 | | Structure of the cystathionine beta-synthase pair domain of the putative Mg2+ transporter SO5017 from Shewanella oneidensis MR-1. | | Descriptor: | CHLORIDE ION, Magnesium transporter, putative | | Authors: | Fratczak, Z, Zimmerman, M.D, Chruszcz, M, Cymborowski, M, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-03 | | Release date: | 2009-12-15 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of the cystathionine beta-synthase pair domain of the putative Mg2+ transporter SO5017 from Shewanella oneidensis MR-1.
To be Published
|
|
1UXO
 
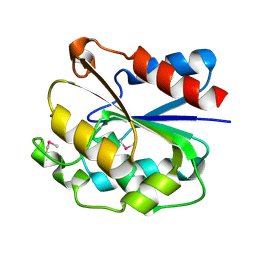 | | The crystal structure of the ydeN gene product from B. subtilis | | Descriptor: | Putative hydrolase YdeN | | Authors: | Janda, I.K, Devedjiev, Y, Cooper, D.R, Chruszcz, M, Derewenda, U, Gabrys, A, Minor, W, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-27 | | Release date: | 2004-05-27 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Harvesting the high-hanging fruit: the structure of the YdeN gene product from Bacillus subtilis at 1.8 angstroms resolution.
Acta Crystallogr. D Biol. Crystallogr., 60, 2004
|
|
3V0R
 
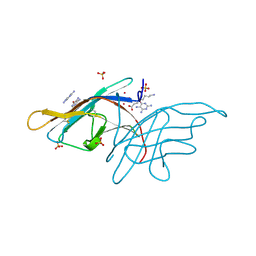 | | Crystal structure of Alternaria alternata allergen Alt a 1 | | Descriptor: | 2,5,6-triaminopyrimidin-4-ol, 8-aminooctanoic acid, Major allergen Alt a 1, ... | | Authors: | Chruszcz, M, Solberg, R, Osinski, T, Chapman, M.D, Minor, W. | | Deposit date: | 2011-12-08 | | Release date: | 2012-06-13 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternaria alternata allergen Alt a 1: a unique beta-barrel protein dimer found exclusively in fungi.
J.Allergy Clin.Immunol., 130, 2012
|
|
1F8N
 
 | |
1FGQ
 
 | |
1FGT
 
 | |
3O4F
 
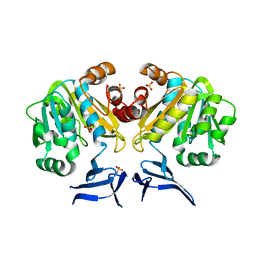 | | Crystal Structure of Spermidine Synthase from E. coli | | Descriptor: | SULFATE ION, Spermidine synthase | | Authors: | Zhou, X, Tkaczuk, K.L, Chruszcz, M, Chua, T.K, Minor, W, Sivaraman, J. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of Escherichia coli spermidine synthase SpeE reveals a unique substrate-binding pocket
J.Struct.Biol., 169, 2010
|
|
