3RQ8
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis soaked with P1,P5-Di(adenosine-5') pentaphosphate | | Descriptor: | ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
5VDB
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with bisubstrate analog 3 | | Descriptor: | (3R,5S,9R,26S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14,20-trioxo-26-({[(phenylacetyl)amino]acetyl}amino)-2,4,6-trioxa-18-thia-11,15,21-triaza-3,5-diphosphaheptacosan-27-oic acid 3,5-dioxide (non-preferred name), SULFATE ION, acetyltransferase PA4794 | | Authors: | Majorek, K.A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-04-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
FEBS Lett., 591, 2017
|
|
5VG6
 
 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 in complex with NADPH and MES. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lipowska, J, Shabalin, I.G, Kutner, J, Gasiorowska, O.A, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-04-10 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 in complex with NADPH and MES.
to be published
|
|
5V0S
 
 | | Crystal structure of the ACT domain of prephenate dehydrogenase tyrA from Bacillus anthracis | | Descriptor: | CALCIUM ION, Prephenate dehydrogenase, SULFATE ION | | Authors: | Shabalin, I.G, Hou, J, Cymborowski, M.T, Otwinowski, Z, Kwon, K, Christendat, D, Gritsunov, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-08 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
5UOG
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in apo form | | Descriptor: | NADPH-dependent glyoxylate/hydroxypyruvate reductase, SULFATE ION | | Authors: | Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Cooper, D.R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-01-31 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5VE2
 
 | | Crystal structure of enoyl-CoA hydratase/isomerase from Pseudoalteromonas atlantica T6c at 2.3 A resolution. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase, GLYCEROL, ... | | Authors: | Siuda, M.K, Shabalin, I.G, Cooper, D.R, Chapman, H.C, Tkaczuk, K.L, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-04-03 | | Release date: | 2017-04-19 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase/isomerase from Pseudoalteromonas atlantica T6c at 2.3 A resolution.
to be published
|
|
2B2W
 
 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
5VGM
 
 | | Crystal structure of dihydroorotase pyrC from Vibrio cholerae in complex with zinc at 1.95 A resolution. | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydroorotase, ... | | Authors: | Lipowska, J, Shabalin, I.G, Miks, C.D, Winsor, J, Cooper, D.R, Shuvalova, L, Kwon, K, Lewinski, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-11 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.
Int.J.Biol.Macromol., 136, 2019
|
|
3S7I
 
 | | Crystal structure of Ara h 1 | | Descriptor: | Allergen Ara h 1, clone P41B, CHLORIDE ION | | Authors: | Chruszcz, M, Maleki, S.J, Solberg, R, Minor, W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Immunologic Characterization of Ara h 1, a Major Peanut Allergen.
J.Biol.Chem., 286, 2011
|
|
2B2T
 
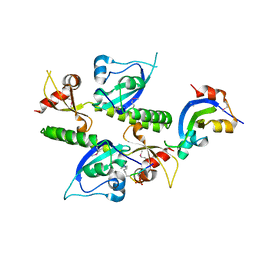 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and phosphothreonine 3 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 tail | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2AZ4
 
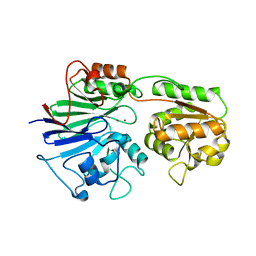 | | Crystal Structure of a Protein of Unknown Function from Enterococcus faecalis V583 | | Descriptor: | ZINC ION, hypothetical protein EF2904 | | Authors: | Zhang, R, Maltseva, N, Moy, S, Collart, F, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-09 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of a hypothetical protein from Enterococcus faecalis V583
To be Published
|
|
5HOZ
 
 | |
2B2Y
 
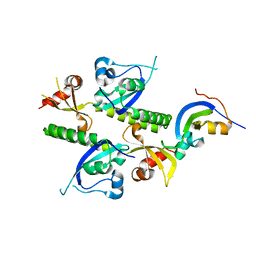 | | Tandem chromodomains of human CHD1 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
5VF2
 
 | | scFv 2D10 re-refined as a complex with trehalose replacing the original alpha-1,6-mannobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, UNKNOWN ATOM OR ION, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
7KPS
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with AcCoA | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL COENZYME *A, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Reidl, C.T, Becker, D.P, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
7KPP
 
 | | Structure of the E102A mutant of a GNAT superfamily PA3944 acetyltransferase | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, COENZYME A, ... | | Authors: | Czub, M.P, Porebski, P.J, Majorek, K.A, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
7KYE
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with CHES | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetyltransferase PA3944, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with CHES
To Be Published
|
|
3RQ2
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis co-crystallized with ATP/Mg2+ and soaked with NADH | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
2B2V
 
 | | Crystal structure analysis of human CHD1 chromodomains 1 and 2 bound to histone H3 resi 1-15 MeK4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2U
 
 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
3UEG
 
 | | Crystal structure of human Survivin K62A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, TETRAETHYLENE GLYCOL, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEC
 
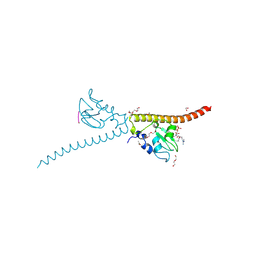 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Baculoviral IAP repeat-containing protein 5, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Cooper, D.R, Chruszcz, M, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEI
 
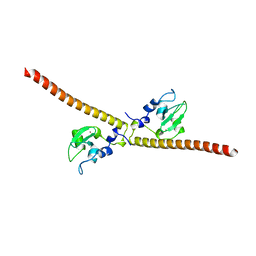 | | Crystal structure of human Survivin E65A mutant | | Descriptor: | Baculoviral IAP repeat-containing protein 5, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
5IIU
 
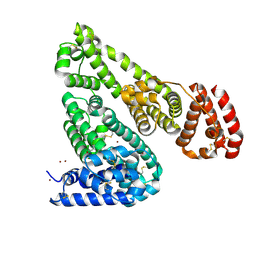 | | Crystal structure of Equine Serum Albumin in the presence of 10 mM zinc at pH 6.9 | | Descriptor: | SULFATE ION, Serum albumin, ZINC ION | | Authors: | Handing, K.B, Shabalin, I.G, Cooper, D.R, Cymborowski, M.T, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
3UEF
 
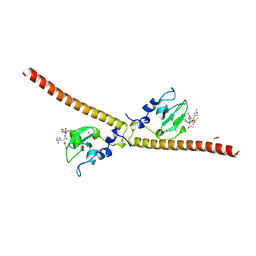 | | Crystal structure of human Survivin bound to histone H3 (C2 space group). | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
