1RTT
 
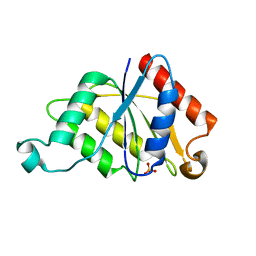 | | Crystal structure determination of a putative NADH-dependent reductase using sulfur anomalous signal | | Descriptor: | SULFATE ION, conserved hypothetical protein | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure determination of an FMN reductase from Pseudomonas aeruginosa PA01 using sulfur anomalous signal.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
3AA0
 
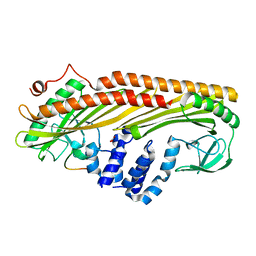 | | Crystal structure of Actin Capping Protein in complex with the Cp-binding motif derived from CARMIL | | Descriptor: | 21mer peptide from Leucine-rich repeat-containing protein 16A, CARBONATE ION, F-actin-capping protein subunit alpha-1, ... | | Authors: | Takeda, S, Minakata, S, Narita, A, Kitazawa, M, Yamakuni, T, Maeda, Y, Nitanai, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two distinct mechanisms for actin capping protein regulation--steric and allosteric inhibition
Plos Biol., 8, 2010
|
|
2G6T
 
 | |
3CUZ
 
 | |
1Y9Q
 
 | |
1YBF
 
 | |
1YDF
 
 | |
2XAJ
 
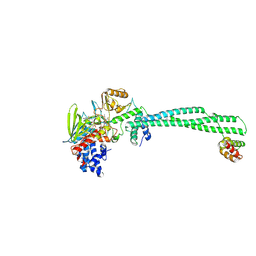 | | Crystal structure of LSD1-CoREST in complex with (-)-trans-2- phenylcyclopropyl-1-amine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, PHENYLETHYLENECARBOXYLIC ACID, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
1RV9
 
 | |
1RRM
 
 | |
1C4F
 
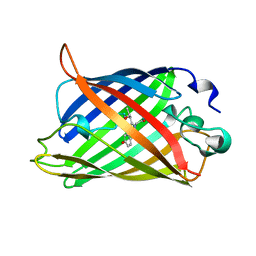 | | GREEN FLUORESCENT PROTEIN S65T AT PH 4.6 | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Elsliger, M.A, Wachter, R.M, Kallio, K, Hanson, G.T, Remington, S.J. | | Deposit date: | 1999-08-21 | | Release date: | 1999-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and spectral response of green fluorescent protein variants to changes in pH.
Biochemistry, 38, 1999
|
|
3MAB
 
 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED PROTEIN FROM LISTERIA MONOCYTOGENES, Triclinic FORM | | Descriptor: | uncharacterized protein | | Authors: | Madegowda, M, Chruszcz, M, Minor, W, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of an uncharacterized protein from listeria monocytogenes
To be Published
|
|
1AL6
 
 | |
1RW0
 
 | |
2AHA
 
 | |
1S0B
 
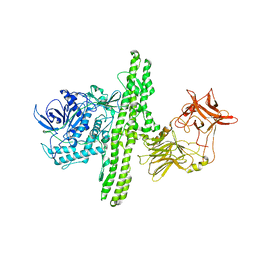 | | Crystal structure of botulinum neurotoxin type B at pH 4.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
2D3E
 
 | |
1Y9H
 
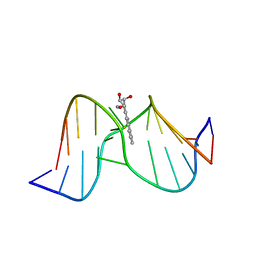 | | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*CP*CP*AP*TP*(5CM)P*(BPG)P*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Zhang, N, Lin, C, Huang, X, Kolbanovskiy, A, Hingerty, B.E, Amin, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-22 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement.
J.Mol.Biol., 346, 2005
|
|
1ZL6
 
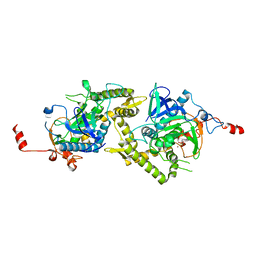 | | Crystal structure of Tyr350Ala mutant of Clostridium botulinum neurotoxin E catalytic domain | | Descriptor: | SULFATE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-05 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
3AA6
 
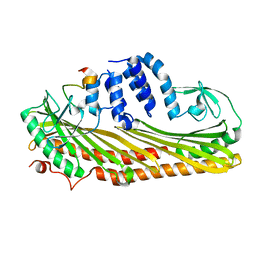 | | Crystal structure of Actin capping protein in complex with the Cp-binding motif derived from CD2AP | | Descriptor: | 23mer peptide from CD2-associated protein, BARIUM ION, F-actin-capping protein subunit alpha-1, ... | | Authors: | Takeda, S, Minakata, S, Narita, A, Kitazawa, M, Yamakuni, T, Maeda, Y, Nitanai, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two distinct mechanisms for actin capping protein regulation--steric and allosteric inhibition
Plos Biol., 8, 2010
|
|
1YV9
 
 | |
1P7T
 
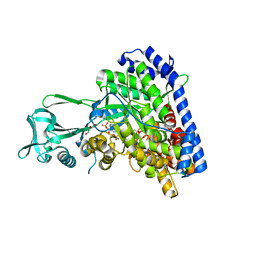 | | Structure of Escherichia coli malate synthase G:pyruvate:acetyl-Coenzyme A abortive ternary complex at 1.95 angstrom resolution | | Descriptor: | ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Anstrom, D.M, Kallio, K, Remington, S.J. | | Deposit date: | 2003-05-05 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Escherichia Coli Malate Synthase G:pyruvate:acetyl-coenzyme A Abortive Ternary Complex at 1.95 Angstrom Resolution
Protein Sci., 12, 2003
|
|
4DLL
 
 | | Crystal structure of a 2-hydroxy-3-oxopropionate reductase from Polaromonas sp. JS666 | | Descriptor: | 2-hydroxy-3-oxopropionate reductase, SULFATE ION | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of a 2-hydroxy-3-oxopropionate reductase from Polaromonas sp. JS666
To be Published
|
|
1BU6
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | GLYCEROL, PROTEIN (GLYCEROL KINASE), SULFATE ION | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
4DVJ
 
 | | Crystal structure of a putative zinc-dependent alcohol dehydrogenase protein from Rhizobium etli CFN 42 | | Descriptor: | Putative zinc-dependent alcohol dehydrogenase protein | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hellerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-23 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of a putative zinc-dependent alcohol dehydrogenase protein from Rhizobium etli CFN 42
To be Published
|
|
