7NUX
 
 | | Crystal structure of the TIR domain of human TLR1 (crystallised without ZN2+ ions) | | Descriptor: | Toll-like receptor 1 | | Authors: | Vakhrameev, D.D, Luginina, A.P, Shevtsov, M.B, Lushpa, V.A, Mineev, K.S, Borshchevskiy, V.I. | | Deposit date: | 2021-03-15 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Modulation of Toll-like receptor 1 intracellular domain structure and activity by Zn 2+ ions.
Commun Biol, 4, 2021
|
|
7AJP
 
 | | Crystal Structure of Human Adenovirus 56 Fiber Knob | | Descriptor: | 1,2-ETHANEDIOL, Fiber, GLYCEROL, ... | | Authors: | Strebl, M, Mindler, K, Stehle, T. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.382018 Å) | | Cite: | Human species D adenovirus hexon capsid protein mediates cell entry through a direct interaction with CD46.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3N95
 
 | |
3K1D
 
 | | Crystal structure of glycogen branching enzyme synonym: 1,4-alpha-D-glucan:1,4-alpha-D-GLUCAN 6-glucosyl-transferase from mycobacterium tuberculosis H37RV | | Descriptor: | 1,4-alpha-glucan-branching enzyme | | Authors: | Pal, K, Kumar, S, Swaminathan, K. | | Deposit date: | 2009-09-27 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of full-length Mycobacterium tuberculosis H37Rv glycogen branching enzyme: insights of N-terminal beta-sandwich in substrate specificity and enzymatic activity
J.Biol.Chem., 285, 2010
|
|
1HI3
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine 2'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
3N93
 
 | | Crystal structure of human CRFR2 alpha extracellular domain in complex with Urocortin 3 | | Descriptor: | GLYCEROL, Maltose binding protein-CRFR2 alpha, Urocortin-3, ... | | Authors: | Pal, K, Swaminathan, K, Pioszak, A.A, Xu, H.E. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of ligand selectivity in human CRFR1 and CRFR2 alpha extracellular domain
To be Published
|
|
5WVX
 
 | | Crystal Structure of bifunctional Kunitz type Trypsin /amylase inhibitor (AMTIN) from the tubers of Alocasia macrorrhiza | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CITRIC ACID, Trypsin/chymotrypsin inhibitor | | Authors: | Palayam, M, Radhakrishnan, M, Lakshminarayanan, K, Balu, K.E, Ganapathy, J, Krishnasamy, G. | | Deposit date: | 2016-12-29 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural insights into a multifunctional inhibitor, 'AMTIN' from tubers of Alocasia macrorrhizos and its possible role in dengue protease (NS2B-NS3) inhibition.
Int. J. Biol. Macromol., 113, 2018
|
|
7W7R
 
 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
7VNX
 
 | | Crystal structure of TkArkI | | Descriptor: | GUANOSINE, TkArkI | | Authors: | Yamashita, S, Minowa, K, Ohira, T, Suzuki, T, Tomita, K. | | Deposit date: | 2021-10-12 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Reversible RNA phosphorylation stabilizes tRNA for cellular thermotolerance.
Nature, 605, 2022
|
|
8AO1
 
 | | solution structure of nanoFAST fluorogen-activating protein in the apo state | | Descriptor: | nanoFAST | | Authors: | Lushpa, V.A, Goncharuk, M.V, Goncharuk, S.A, Baranov, M.S, Mineev, K.S. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of NanoFAST in the Apo State and in Complex with its Fluorogen HBR-DOM2.
Int J Mol Sci, 23, 2022
|
|
8AO0
 
 | | Solution structure of nanoFAST/HBR-DOM2 complex | | Descriptor: | (5~{Z})-5-[(2,5-dimethoxy-4-oxidanyl-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Photoactive yellow protein | | Authors: | Lushpa, V.A, Goncharuk, M.V, Goncharuk, S.A, Baleeva, N.S, Baranov, M.S, Mineev, K.S. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of NanoFAST in the Apo State and in Complex with its Fluorogen HBR-DOM2.
Int J Mol Sci, 23, 2022
|
|
3MFK
 
 | |
6LKB
 
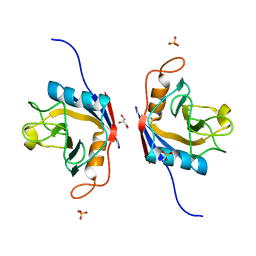 | | Crystal Structure of the peptidylprolyl isomerase domain of Arabidopsis thaliana CYP71. | | Descriptor: | COBALT (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lakhanpal, S, Jobichen, C, Swaminathan, K. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analyses of the PPIase domain of Arabidopsis thaliana CYP71 reveal its catalytic activity toward histone H3.
Febs Lett., 595, 2021
|
|
7AV6
 
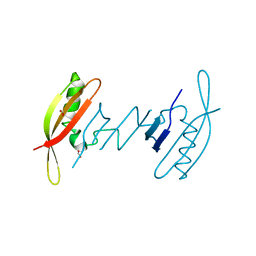 | | FAST in a domain-swapped dimer form | | Descriptor: | FORMIC ACID, Photoactive yellow protein | | Authors: | Bukhdruker, S, Remeeva, A, Ruchkin, D, Gorbachev, D, Povarova, N, Mineev, K, Goncharuk, S, Baranov, M, Mishin, A, Borshchevskiy, V. | | Deposit date: | 2020-11-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NanoFAST: structure-based design of a small fluorogen-activating protein with only 98 amino acids.
Chem Sci, 12, 2021
|
|
8OYD
 
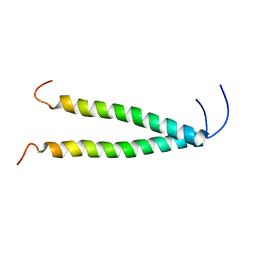 | |
8AR3
 
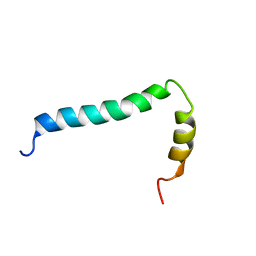 | | Solution structure of TLR9 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 9 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR0
 
 | | Solution structure of TLR2 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 2 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
3OQT
 
 | | Crystal structure of Rv1498A protein from mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, Rv1498A PROTEIN, SODIUM ION | | Authors: | Liu, F, Xiong, J, Kumar, S, Yang, C, Li, S, Ge, S, Xia, N, Swaminathan, K. | | Deposit date: | 2010-09-04 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural and biophysical characterization of Mycobacterium tuberculosis dodecin Rv1498A.
J.Struct.Biol., 175, 2011
|
|
8AR2
 
 | | Solution structure of TLR5 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 5 | | Authors: | Shabalkina, A.V, Kornilov, F.D, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
6HJ7
 
 | |
8AR1
 
 | | Solution structure of TLR3 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 3 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8UZW
 
 | |
3K9Z
 
 | | Rational Design of a Structural and Functional Nitric Oxide Reductase | | Descriptor: | FE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yeung, N, Lin, Y.-W, Gao, Y.-G, Zhao, X, Russell, B.S, Lei, L, Miner, K.D, Robinson, H, Lu, Y. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of a structural and functional nitric oxide reductase.
Nature, 462, 2009
|
|
7W7S
 
 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
7VTJ
 
 | | The cross-reaction complex structure with VQIIYK peptide and tau antibody's Fab domain. | | Descriptor: | Heavy chain of Fab, Light chain of Fab, VQIIYK peptide | | Authors: | Tsuchida, T, Fukuhara, N, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, Y, Ishida, T, Tomoo, K. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The cross-reaction complex structure with VQIIYK peptide and tau antibody's Fab domain.
To Be Published
|
|
