2P0K
 
 | | Crystal structure of SCMH1 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Polycomb protein SCMH1 | | Authors: | Herzanych, N, Senisterra, G, Liu, Y, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of SCMH1
To be Published
|
|
2P0W
 
 | | Human histone acetyltransferase 1 (HAT1) | | Descriptor: | ACETAMIDE, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Wu, H, Min, J, Zeng, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human histone acetyltransferase 1 (HAT1) in complex with acetylcoenzyme A and histone peptide H4
To be Published
|
|
6B57
 
 | | tudor in complex with ligand | | Descriptor: | Tudor and KH domain-containing protein, UNKNOWN ATOM OR ION | | Authors: | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2LLK
 
 | | Solution NMR structure of the N-terminal myb-like 1 domain of the human cyclin-D-binding transcription factor 1 (hDMP1), Northeast Structural Genomics Consortium (NESG) target ID hr8011a | | Descriptor: | Cyclin-D-binding Myb-like transcription factor 1 | | Authors: | Montecchio, M, Lemak, A, Yee, A, Xu, C, Garcia, M, Houliston, S, Min, J, Bellanda, M, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-10 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the N-terminal myb-like 1 domain of the human cyclin D binding transcription factor 1 (hDMP1).
To be Published
|
|
2LLL
 
 | | Solution NMR structure of C-terminal globular domain of human Lamin-B2, Northeast Structural Genomics Consortium target HR8546A | | Descriptor: | Lamin-B2 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Xu, C, Min, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | Deposit date: | 2011-11-11 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of c-terminal globular domain of human Lamin-B2
To be Published
|
|
4YVD
 
 | | Crytsal structure of human Pleiotropic Regulator 1 (PRL1) | | Descriptor: | CHLORIDE ION, Pleiotropic regulator 1, SODIUM ION, ... | | Authors: | Dong, A, Zeng, H, Xu, C, Tempel, W, Li, Y, He, H, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crytsal structure of human Pleiotropic Regulator 1 (PRL1).
to be published
|
|
2LV9
 
 | | Solution NMR structure of the PHD domain of human MLL5, Northeast structural genomics consortium target HR6512A | | Descriptor: | Histone-lysine N-methyltransferase MLL5, ZINC ION | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Wu, H, Min, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | Deposit date: | 2012-06-29 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the human MLL5 PHD domain (CASP Target)
To be Published
|
|
4Z3C
 
 | | Zinc finger region of human TET3 in complex with CpG DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*CP*GP*TP*TP*GP*GP*C)-3'), Methylcytosine dioxygenase, UNKNOWN ATOM OR ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Dong, A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-31 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 2017
|
|
6CDG
 
 | | GID4 fragment in complex with a peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Hexapeptide PGLWKS, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CD8
 
 | | Complex of GID4 fragment with short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PSRV, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CDC
 
 | | GID4 in complex with a tetrapeptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PGLW, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CCT
 
 | | Fragment of GID4 in complex with a short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6D1T
 
 | | Complex of MBD1-MBD and methylated DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 1, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Complex of MBD1-MBD and methylated DNA
to be published
|
|
5HZM
 
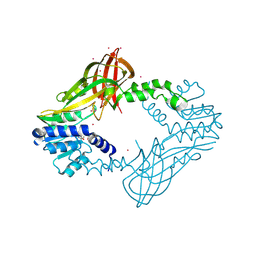 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN ATOM OR ION | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, MIN, J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-02 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
6CCR
 
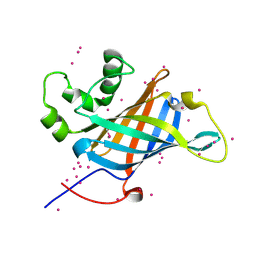 | | Selenomethionyl derivative of a GID4 fragment | | Descriptor: | Glucose-induced degradation protein 4 homolog, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-04-04 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
4Z0O
 
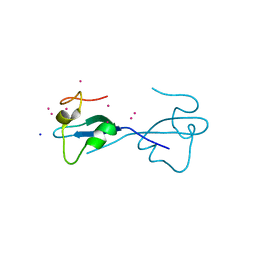 | | CW-type zinc finger of ZCWPW2 with F78D mutation | | Descriptor: | SODIUM ION, UNKNOWN ATOM OR ION, ZINC ION, ... | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | CW-type zinc finger of ZCWPW2 with F78D mutation
To be Published
|
|
4Z0R
 
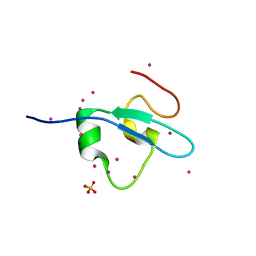 | | Crystal Structure of the CW domain of ZCWPW2 mutant F78R in complex with histone H3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Histone H3.1, SULFATE ION, ... | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the CW domain of ZCWPW2 mutant F78R in complex with histone H3 peptide
To be Published
|
|
6CCU
 
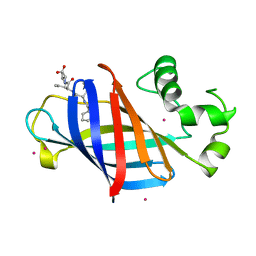 | | Complex between a GID4 fragment and a short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Short peptide, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CD9
 
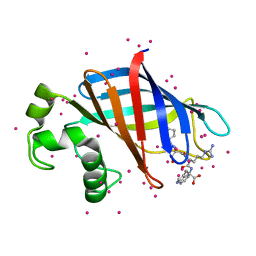 | | GID4 in complex with a peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PSRW, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6DUB
 
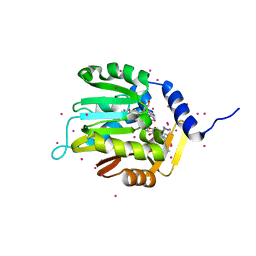 | | Crystal structure of a methyltransferase | | Descriptor: | Alpha N-terminal protein methyltransferase 1B, GLYCEROL, RCC1, ... | | Authors: | Dong, C, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An asparagine/glycine switch governs product specificity of human N-terminal methyltransferase NTMT2.
Commun Biol, 1, 2018
|
|
6C1Y
 
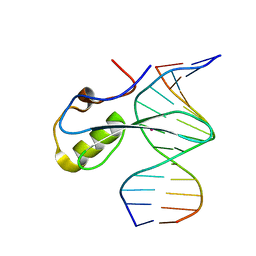 | | mbd of human mecp2 in complex with methylated DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Bian, C, Tempel, W, Wernimont, A.K, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
6C2F
 
 | | MBD2 in complex with methylated DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-08 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | MBD2 in complex with methylated DNA
to be published
|
|
3K35
 
 | | Crystal Structure of Human SIRT6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
2L12
 
 | | Solution NMR structure of the chromobox protein 7 with H3K9me3 | | Descriptor: | Chromobox homolog 7, Histone H3 | | Authors: | Kaustov, L, Lemak, A, Gutmanas, A, Fares, C, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2L11
 
 | | Solution NMR structure of the Cbx3 in complex with H3K9me3 peptide | | Descriptor: | Chromobox protein homolog 3, Histone H3 | | Authors: | Kaustov, L, Lemak, A, Fares, C, Gutmanas, A, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
