1Z1F
 
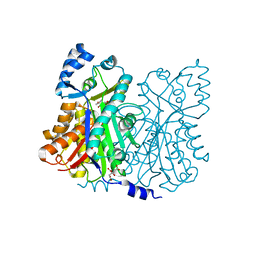 | | Crystal structure of stilbene synthase from Arachis hypogaea (resveratrol-bound form) | | Descriptor: | CITRIC ACID, RESVERATROL, stilbene synthase | | Authors: | Shomura, Y, Torayama, I, Suh, D.Y, Xiang, T, Kita, A, Sankawa, U, Miki, K. | | Deposit date: | 2005-03-03 | | Release date: | 2005-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of stilbene synthase from Arachis hypogaea
Proteins, 60, 2005
|
|
1Z1E
 
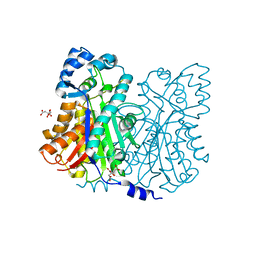 | | Crystal structure of stilbene synthase from Arachis hypogaea | | Descriptor: | CITRIC ACID, stilbene synthase | | Authors: | Shomura, Y, Torayama, I, Suh, D.Y, Xiang, T, Kita, A, Sankawa, U, Miki, K. | | Deposit date: | 2005-03-03 | | Release date: | 2005-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of stilbene synthase from Arachis hypogaea
Proteins, 60, 2005
|
|
6JBD
 
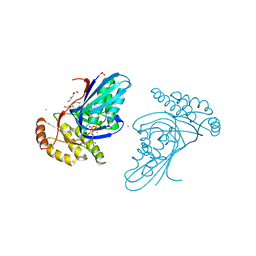 | | Phosphotransferase-ATP complex related to CoA biosynthesis pathway | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kita, A, Kishimoto, A, Shimosaka, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of pantoate kinase from Thermococcus kodakarensis.
Proteins, 88, 2020
|
|
7E99
 
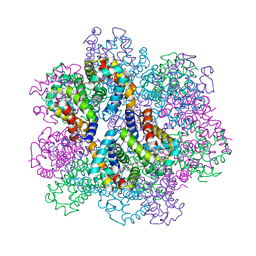 | | Oxy-deoxy intermediate of 400 kDa giant hemoglobin at 13% oxygen saturation | | Descriptor: | Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, Extracellular giant hemoglobin major globin subunit B2, ... | | Authors: | Numoto, N, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Coarse snapshots of oxygen-dissociation intermediates of a giant hemoglobin elucidated by determining the oxygen saturation in individual subunits in the crystalline state.
Iucrj, 8, 2021
|
|
7E97
 
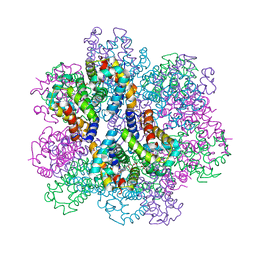 | | Oxy-deoxy intermediate of 400 kDa giant hemoglobin at 58% oxygen saturation | | Descriptor: | Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, Extracellular giant hemoglobin major globin subunit B2, ... | | Authors: | Numoto, N, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Coarse snapshots of oxygen-dissociation intermediates of a giant hemoglobin elucidated by determining the oxygen saturation in individual subunits in the crystalline state.
Iucrj, 8, 2021
|
|
7E98
 
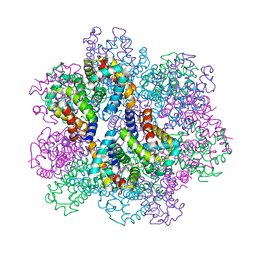 | | Oxy-deoxy intermediate of 400 kDa giant hemoglobin at 21% oxygen saturation | | Descriptor: | Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, Extracellular giant hemoglobin major globin subunit B2, ... | | Authors: | Numoto, N, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Coarse snapshots of oxygen-dissociation intermediates of a giant hemoglobin elucidated by determining the oxygen saturation in individual subunits in the crystalline state.
Iucrj, 8, 2021
|
|
7E96
 
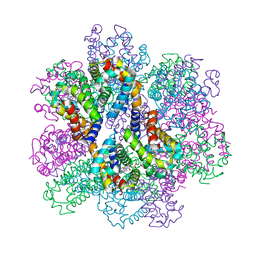 | | Oxy-deoxy intermediate of 400 kDa giant hemoglobin at 69% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, ... | | Authors: | Numoto, N, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Coarse snapshots of oxygen-dissociation intermediates of a giant hemoglobin elucidated by determining the oxygen saturation in individual subunits in the crystalline state.
Iucrj, 8, 2021
|
|
7E4L
 
 | |
6JBC
 
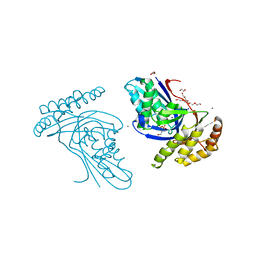 | | Phosphotransferase related to CoA biosynthesis pathway | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Kita, A, Kishimoto, A, Shimosaka, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pantoate kinase from Thermococcus kodakarensis.
Proteins, 88, 2020
|
|
1OWN
 
 | | DATA3:DNA photolyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWP
 
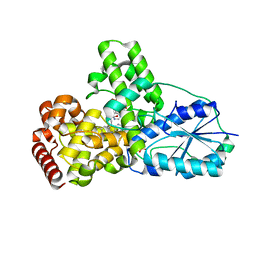 | | DATA6:photoreduced DNA pholyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWO
 
 | | DATA4:photoreduced DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWM
 
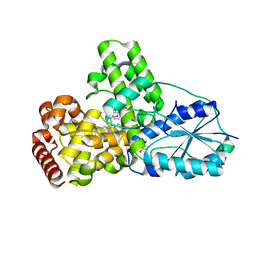 | | DATA1:DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1REP
 
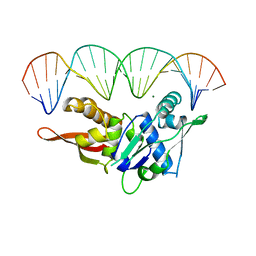 | | CRYSTAL STRUCTURE OF REPLICATION INITIATOR PROTEIN REPE54 OF MINI-F PLASMID COMPLEXED WITH AN ITERON DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*T)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Komori, H, Matsunaga, F, Higuchi, Y, Ishiai, M, Wada, C, Miki, K. | | Deposit date: | 1999-04-29 | | Release date: | 2000-02-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6 A resolution.
EMBO J., 18, 1999
|
|
5AYV
 
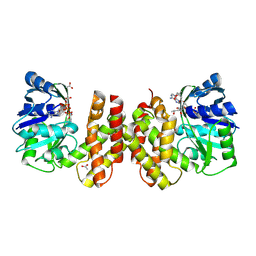 | | Crystal structure of archaeal ketopantoate reductase complexed with coenzyme A and 2-oxopantoate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-dehydropantoate 2-reductase, ACETATE ION, ... | | Authors: | Aikawa, Y, Nishitani, Y, Miki, K. | | Deposit date: | 2015-09-08 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal structure of archaeal ketopantoate reductase complexed with coenzyme a and 2-oxopantoate provides structural insights into feedback regulation
Proteins, 84, 2016
|
|
1OWL
 
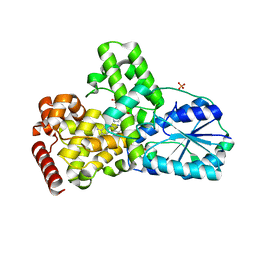 | | Structure of apophotolyase from Anacystis nidulans | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PRC
 
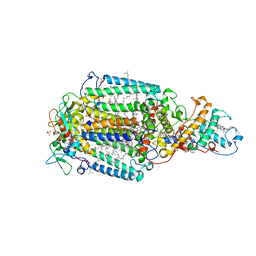 | | CRYSTALLOGRAPHIC REFINEMENT AT 2.3 ANGSTROMS RESOLUTION AND REFINED MODEL OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS | | Descriptor: | 15-trans-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Deisenhofer, J, Epp, O, Miki, K, Huber, R, Michel, H. | | Deposit date: | 1988-02-04 | | Release date: | 1989-01-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic refinement at 2.3 A resolution and refined model of the photosynthetic reaction centre from Rhodopseudomonas viridis.
J.Mol.Biol., 246, 1995
|
|
6A2J
 
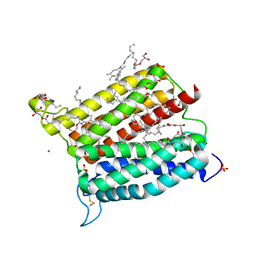 | | Crystal structure of heme A synthase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Heme A synthase, ... | | Authors: | Niwa, S, Takeda, K, Kosugi, M, Tsutsumi, E, Miki, K. | | Deposit date: | 2018-06-12 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of heme A synthase fromBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AIR
 
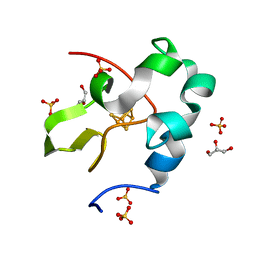 | | High resolution structure of perdeuterated high-potential iron-sulfur protein | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-08-24 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Characterization of perdeuterated high-potential iron-sulfur protein with high-resolution X-ray crystallography.
Proteins, 88, 2020
|
|
5ZIL
 
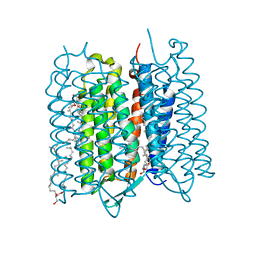 | | Crystal structure of bacteriorhodopsin at 1.29 A resolution | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Hasegawa, N, Jonotsuka, H, Miki, K, Takeda, K. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | X-ray structure analysis of bacteriorhodopsin at 1.3 angstrom resolution.
Sci Rep, 8, 2018
|
|
5ZIM
 
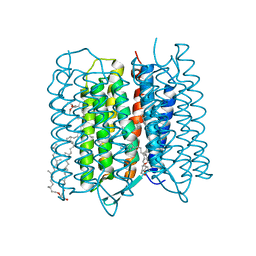 | | Crystal structure of bacteriorhodopsin at 1.25 A resolution | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Hasegawa, N, Jonotsuka, H, Miki, K, Takeda, K. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | X-ray structure analysis of bacteriorhodopsin at 1.3 angstrom resolution.
Sci Rep, 8, 2018
|
|
5ZCA
 
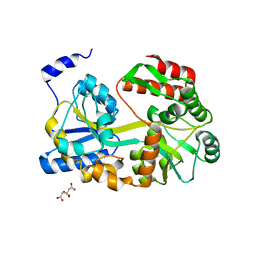 | | Crystal structure of lambda repressor (1-20) fused with maltose-binding protein | | Descriptor: | CITRIC ACID, Repressor protein cI,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-02-16 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Co-translational folding of alpha-helical proteins: structural studies of intermediate-length variants of the lambda repressor
Febs Open Bio, 8, 2018
|
|
5ZIN
 
 | | Crystal structure of bacteriorhodopsin at 1.27 A resolution | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Hasegawa, N, Jonotsuka, H, Miki, K, Takeda, K. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | X-ray structure analysis of bacteriorhodopsin at 1.3 angstrom resolution.
Sci Rep, 8, 2018
|
|
6AIQ
 
 | | High resolution structure of recombinant high-potential iron-sulfur protein | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-08-24 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Characterization of perdeuterated high-potential iron-sulfur protein with high-resolution X-ray crystallography.
Proteins, 88, 2020
|
|
1UD6
 
 | | Crystal structure of AmyK38 with potassium ion | | Descriptor: | POTASSIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
