2DKA
 
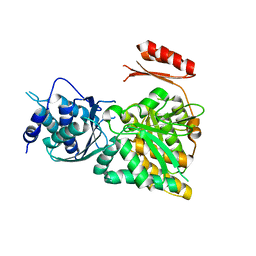 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the apo-form | | Descriptor: | Phosphoacetylglucosamine mutase | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
2ED4
 
 | | Crystal structure of flavin reductase HpaC complexed with FAD and NAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygenae | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2E6E
 
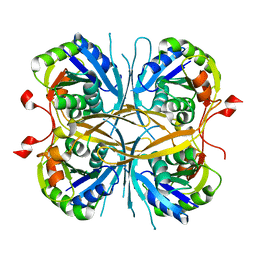 | |
1V9M
 
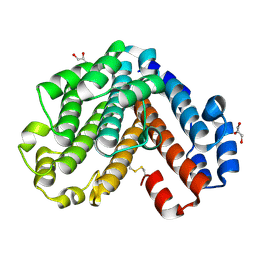 | | Crystal structure of the C subunit of V-type ATPase from Thermus thermophilus | | Descriptor: | GLYCEROL, V-type ATP synthase subunit C | | Authors: | Numoto, N, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the C subunit of V-type ATPase from Thermus thermophilus at 1.85 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2D2F
 
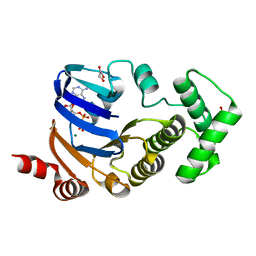 | | Crystal structure of atypical cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Watanabe, S, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-09-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Atypical Cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8.
J.Mol.Biol., 353, 2005
|
|
2ECU
 
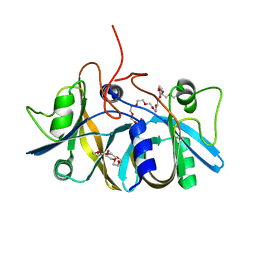 | | Crystal structure of flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, DODECAETHYLENE GLYCOL, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygnease | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2YYJ
 
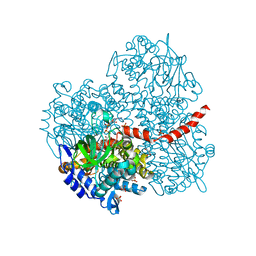 | | Crystal structure of the oxygenase component (HpaB) of 4-hydroxyphenylacetate 3-monooxygenase complexed with FAD and 4-hydroxyphenylacetate | | Descriptor: | 4-HYDROXYPHENYLACETATE, 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYI
 
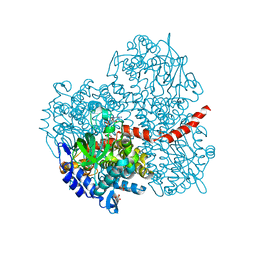 | | Crystal structure of the oxygenase component (HpaB) of 4-hydroxyphenylacetate 3-monooxygenase complexed with FAD | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYG
 
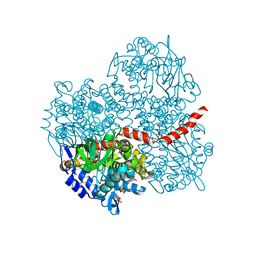 | | Crystal structure of the oxygenase component (HpaB) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, SULFATE ION | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYM
 
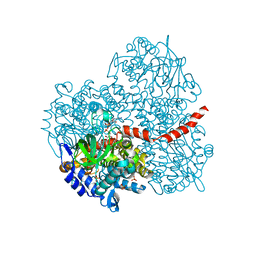 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) complexed with FAD and 4-hydroxyphenylacetate | | Descriptor: | 4-HYDROXYPHENYLACETATE, 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYK
 
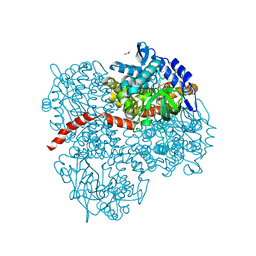 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYL
 
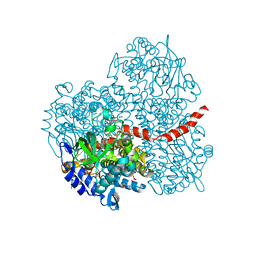 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) complexed with FAD | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
3A38
 
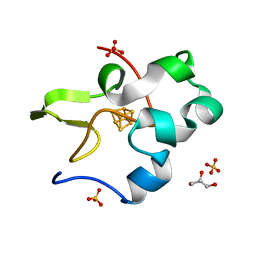 | | Crystal structure of high-potential iron-sulfur protein from Thermochromatium tepidum at 0.7 angstrom resolution | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Takeda, K, Kusumoto, K, Hirano, Y, Miki, K. | | Deposit date: | 2009-06-10 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.7 Å) | | Cite: | Detailed assessment of X-ray induced structural perturbation in a crystalline state protein.
J.Struct.Biol., 169, 2010
|
|
3A5C
 
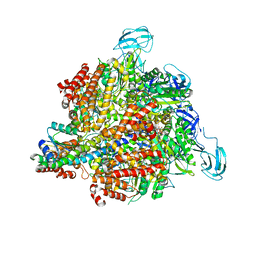 | | Inter-subunit interaction and quaternary rearrangement defined by the central stalk of prokaryotic V1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Numoto, N, Hasegawa, Y, Takeda, K, Miki, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.51 Å) | | Cite: | Inter-subunit interaction and quaternary rearrangement defined by the central stalk of prokaryotic V1-ATPase
Embo Rep., 10, 2009
|
|
3A39
 
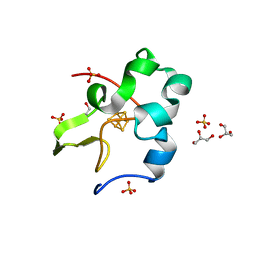 | | Crystal Structure of High-Potential Iron-Sulfur Protein from Thermochromatium tepidum at 0.72 angstrom resolution | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Takeda, K, Kusumoto, K, Hirano, Y, Miki, K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.72 Å) | | Cite: | Detailed assessment of X-ray induced structural perturbation in a crystalline state protein.
J.Struct.Biol., 169, 2010
|
|
2Z4I
 
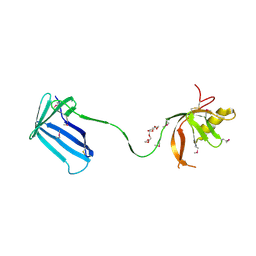 | | Crystal structure of the Cpx pathway activator NlpE from Escherichia coli | | Descriptor: | Copper homeostasis protein cutF, HEXAETHYLENE GLYCOL, SULFATE ION | | Authors: | Hirano, Y, Hossain, M.M, Takeda, K, Tokuda, H, Miki, K. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of the Cpx Pathway Activator NlpE on the Outer Membrane of Escherichia coli
Structure, 15, 2007
|
|
2Z6W
 
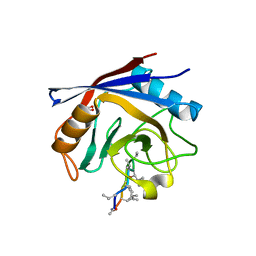 | | Crystal structure of human cyclophilin D in complex with cyclosporin A | | Descriptor: | CITRIC ACID, CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Kajitani, K, Fujihashi, M, Kobayashi, Y, Shimizu, S, Tsujimoto, Y, Miki, K. | | Deposit date: | 2007-08-09 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Crystal Structure of Human Cyclophilin D in Complex with its Inhibitor, Cyclosporin a at 0.96-A Resolution.
Proteins, 70, 2008
|
|
2ZPC
 
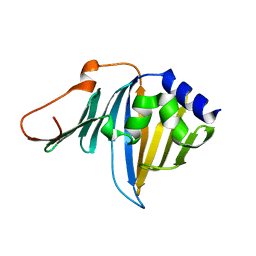 | | Crystal structure of the R43L mutant of LolA in the closed form | | Descriptor: | Outer-membrane lipoprotein carrier protein | | Authors: | Takeda, K, Yokota, N, Oguchi, Y, Tokuda, H, Miki, K. | | Deposit date: | 2008-07-10 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Opening and closing of the hydrophobic cavity of LolA coupled to lipoprotein binding and release.
J.Biol.Chem., 283, 2008
|
|
2ZC8
 
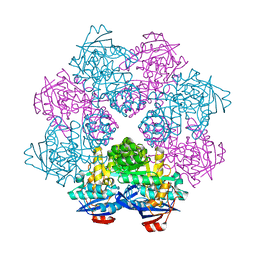 | | Crystal structure of N-Acylamino Acid Racemase from Thermus thermophilus HB8 | | Descriptor: | N-acylamino acid racemase | | Authors: | Hayashida, M, Kim, S.H, Takeda, K, Hisano, T, Miki, K. | | Deposit date: | 2007-11-05 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of N-acylamino acid racemase from Thermus thermophilus HB8
Proteins, 71, 2008
|
|
2ZHH
 
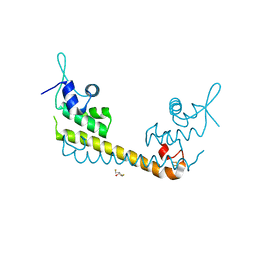 | | Crystal structure of SoxR | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FE2/S2 (INORGANIC) CLUSTER, Redox-sensitive transcriptional activator soxR | | Authors: | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | Deposit date: | 2008-02-05 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZZV
 
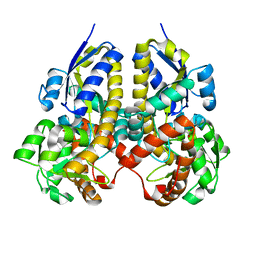 | | Crystal Structure of a Periplasmic Substrate Binding Protein in Complex with Calcium and Lactate | | Descriptor: | ABC transporter, solute-binding protein, CALCIUM ION, ... | | Authors: | Akiyama, N, Takeda, K, Miki, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a periplasmic substrate-binding protein in complex with calcium lactate
J.Mol.Biol., 392, 2009
|
|
2ZZX
 
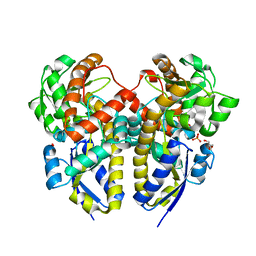 | | Crystal Structure of a Periplasmic Substrate Binding Protein in Complex with Lactate | | Descriptor: | ABC transporter, solute-binding protein, CALCIUM ION, ... | | Authors: | Akiyama, N, Takeda, K, Miki, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a periplasmic substrate-binding protein in complex with calcium lactate
J.Mol.Biol., 392, 2009
|
|
3A12
 
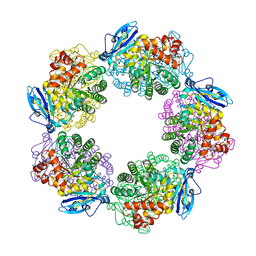 | | Crystal structure of Type III Rubisco complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
3A6M
 
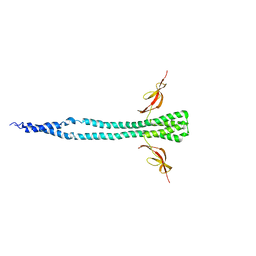 | |
2ZZW
 
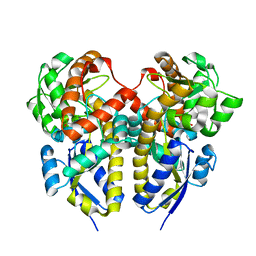 | | Crystal Structure of a Periplasmic Substrate Binding Protein in Complex with Zinc and Lactate | | Descriptor: | ABC transporter, solute-binding protein, LACTIC ACID, ... | | Authors: | Akiyama, N, Takeda, K, Miki, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a periplasmic substrate-binding protein in complex with calcium lactate
J.Mol.Biol., 392, 2009
|
|
