6UR7
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-10-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
4RCK
 
 | |
4RJ0
 
 | |
4RM7
 
 | |
4S1A
 
 | | Crystal structure of a hypothetical protein Cthe_0052 from Ruminiclostridium thermocellum ATCC 27405 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRATE ANION, TETRAETHYLENE GLYCOL, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-09 | | Release date: | 2015-01-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a hypothetical protein Cthe_0052 from Ruminiclostridium thermocellum ATCC27405
To be Published
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RUW
 
 | |
4RPC
 
 | | Crystal structure of the putative alpha/beta hydrolase family protein from Desulfitobacterium hafniense | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TETRAETHYLENE GLYCOL, putative alpha/beta hydrolase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the putative alpha/beta hydrolase family protein from Desulfitobacterium hafniense
To be Published
|
|
4XRR
 
 | | Crystal structure of cals8 from micromonospora echinospora (P294S mutant) | | Descriptor: | CalS8, GLYCEROL | | Authors: | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Characterization of CalS8, a TDP-alpha-D-Glucose Dehydrogenase Involved in Calicheamicin Aminodideoxypentose Biosynthesis.
J. Biol. Chem., 290, 2015
|
|
4DQD
 
 | | The crystal structure of a transporter in complex with 3-phenylpyruvic acid | | Descriptor: | 3-HYDROXYPYRUVIC ACID, 3-PHENYLPYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
5T2X
 
 | | Crystal structure of Uncharacterised protein lpg1670 | | Descriptor: | Uncharacterized protein LPG1670 | | Authors: | Chang, C, Xu, X, Cui, H, SAVCHENKO, A, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-08-24 | | Release date: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Crystal structure of Uncharacterised protein lpg1670
To Be Published
|
|
6UUK
 
 | | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes | | Descriptor: | Muramoyltetrapeptide carboxypeptidase | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-10-30 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes
To Be Published
|
|
4WY2
 
 | | Crystal structure of universal stress protein E from Proteus mirabilis in complex with UDP-3-O-[(3R)-3-hydroxytetradecanoyl]-N-acetyl-alpha-glucosamine | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Shumilin, I.A, Shabalin, I.G, Handing, K.B, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-15 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of universal stress protein E from Proteus mirabilis incomplex withUDP-3-O-[(3R)-3-hydroxytetradecanoyl]-N-acetyl-alpha-glucosamine
to be published
|
|
6W1I
 
 | | Re-interpretation of ppGpp (G4P) electron density in the deposited crystal structure of Xanthine phosphoribosyltransferase (XPRT) (1Y0B). | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, SODIUM ION, Xanthine phosphoribosyltransferase | | Authors: | Satyshur, K.A, Anderson, B.W, Keck, J.L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism of Regulation of the Purine Salvage Enzyme XPRT by the Alarmones pppGpp, ppGpp, and pGpp.
J.Mol.Biol., 432, 2020
|
|
4XA9
 
 | | Crystal structure of the complex between the N-terminal domain of RavJ and LegL1 from Legionella pneumophila str. Philadelphia | | Descriptor: | Gala protein type 1, 3 or 4, Uncharacterized protein | | Authors: | Stogios, P.J, Cuff, M.E, Nocek, B, Evdokimova, E, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-13 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol. Syst. Biol., 12, 2016
|
|
4U4E
 
 | |
5TF0
 
 | | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase family 3 N-terminal domain protein, MAGNESIUM ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-23 | | Release date: | 2016-10-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.2021 Å) | | Cite: | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis
To Be Published, 2016
|
|
7SF2
 
 | | Crystal Structure of Beta-Galactosidase from Bacteroides cellulosilyticus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-10-02 | | Release date: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Beta-Galactosidase from Bacteroides cellulosilyticus
To Be Published
|
|
4X2R
 
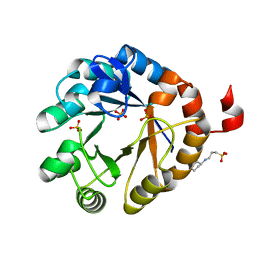 | | Crystal structure of PriA from Actinomyces urogenitalis | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, PHOSPHATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-26 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Evolution of substrate specificity in a retained enzyme driven by gene loss.
Elife, 6, 2017
|
|
6NTR
 
 | | Crystal Structure of Beta-barrel-like Protein of Domain of Unknown Function DUF1849 from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ATP/GTP-binding site-containing protein A, GLYCEROL | | Authors: | Kim, Y, Bigelow, L, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-01-30 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | BrucellaPeriplasmic Protein EipB Is a Molecular Determinant of Cell Envelope Integrity and Virulence.
J.Bacteriol., 201, 2019
|
|
4XR9
 
 | | Crystal structure of CalS8 from Micromonospora echinospora cocrystallized with NAD and TDP-glucose | | Descriptor: | CalS8, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CalS8 from Micromonospora echinospora
To Be Published
|
|
4WUI
 
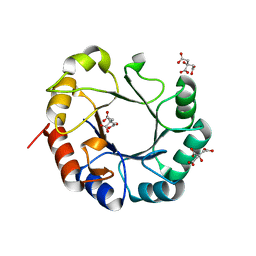 | | Crystal structure of TrpF from Jonesia denitrificans | | Descriptor: | CITRIC ACID, N-(5'-phosphoribosyl)anthranilate isomerase | | Authors: | Michalska, K, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-31 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4WZ2
 
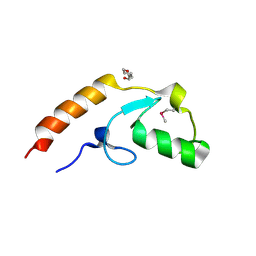 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, Ile175Met mutant | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, HEXANE-1,6-DIOL | | Authors: | Stogios, P.J, Qualie, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-28 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
4WRP
 
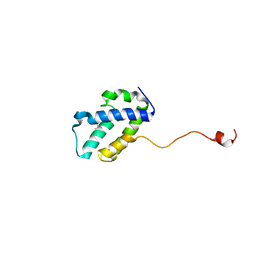 | | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Uncharacterized protein | | Authors: | Cuff, M.E, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-24 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To Be Published
|
|
4X9S
 
 | | CRYSTAL STRUCTURE OF HISAP FROM STREPTOMYCES SP. MG1 | | Descriptor: | Phosphoribosyl isomerase A, SULFATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
