2GEZ
 
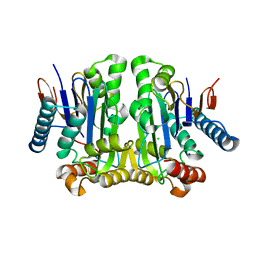 | | Crystal structure of potassium-independent plant asparaginase | | Descriptor: | CHLORIDE ION, L-asparaginase alpha subunit, L-asparaginase beta subunit, ... | | Authors: | Michalska, K, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of plant asparaginase.
J.Mol.Biol., 360, 2006
|
|
3C17
 
 | |
7M5F
 
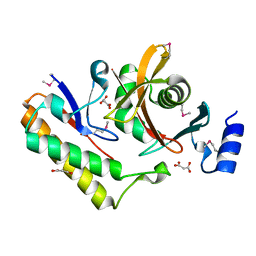 | | Contact-dependent inhibition system from Serratia marcescens BWH57 | | Descriptor: | CdiI, MALONATE ION, Toxin CdiA | | Authors: | Michalska, K, Nutt, W, Stols, L, Jedrzejczak, R, Hayes, C.S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-03-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Contact-dependent inhibition system from Serratia marcescens
To Be Published
|
|
6QKY
 
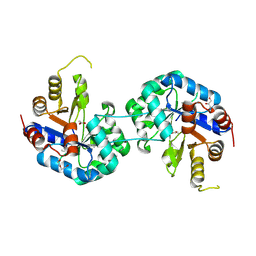 | | Tryptophan synthase subunit alpha from Streptococcus pneumoniae with 3D domain swap in the core of TIM barrel | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Michalska, K, Kowiel, M, Bigelow, L, Endres, M, Gilski, M, Jaskolski, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-27 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | 3D domain swapping in the TIM barrel of the alpha subunit of Streptococcus pneumoniae tryptophan synthase.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2ZAK
 
 | | Orthorhombic crystal structure of precursor E. coli isoaspartyl peptidase/L-asparaginase (EcAIII) with active-site T179A mutation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, L-asparaginase precursor, ... | | Authors: | Michalska, K, Hernandez-Santoyo, A, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal packing of plant-type L-asparaginase from Escherichia coli
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6AZY
 
 | | Crystal structure of Hsp104 R328M/R757M mutant from Calcarisporiella thermophila | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein Hsp104 | | Authors: | Michalska, K, Bigelow, L, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-09-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
3IE5
 
 | | Crystal structure of Hyp-1 protein from Hypericum perforatum (St John's wort) involved in hypericin biosynthesis | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Michalska, K, Fernandes, H, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Crystal structure of Hyp-1, a St. John's wort protein implicated in the biosynthesis of hypericin
J.Struct.Biol., 169, 2010
|
|
5T86
 
 | | Crystal structure of CDI complex from E. coli A0 34/86 | | Descriptor: | ACETATE ION, CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of CDI complex from E. coli A0 34/86
To Be Published
|
|
4DIM
 
 | |
6U8J
 
 | | Crystal structure of 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase/phospho-2-dehydro-3-deoxyheptonate aldolase (Aro3) from Candida auris | | Descriptor: | Phospho-2-dehydro-3-deoxyheptonate aldolase, UNKNOWN ATOM OR ION | | Authors: | Michalska, K, Evdokimova, E, Semper, C, Di Leo, R, Stogios, P.J, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Crystal structure of 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase/phospho-2-dehydro-3-deoxyheptonate aldolase (Aro3) from
Candida auris
To Be Published
|
|
2ZAL
 
 | | Crystal structure of E. coli isoaspartyl aminopeptidase/L-asparaginase in complex with L-aspartate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ASPARTIC ACID, CALCIUM ION, ... | | Authors: | Michalska, K, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate
J.Biol.Chem., 280, 2005
|
|
6U83
 
 | | OmpA-like domain of FopA1 from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-ALANINE, Outer membrane associated protein, ... | | Authors: | Michalska, K, Skarina, T, Stogios, P.J, Di Leo, R, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-04 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3566 Å) | | Cite: | OmpA-like domain of FopA1 from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
6CP8
 
 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CdiA, CdiI, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
6CP9
 
 | | Contact-dependent growth inhibition toxin - immunity protein complex from Klebsiella pneumoniae 342 | | Descriptor: | CdiA, CdiI | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
4YCS
 
 | | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment) | | Descriptor: | ACETATE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Michalska, K, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment)
To Be Published
|
|
4ZXW
 
 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (complex with (R)-(-)-1-(2-naphthyl)-1,2-ethanediol and sucrose) | | Descriptor: | (1R)-1-(naphthalen-2-yl)ethane-1,2-diol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-domain type II peptide synthetase, ... | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus
To Be Published
|
|
4ZNM
 
 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form) | | Descriptor: | C-domain type II peptide synthetase, CHLORIDE ION, SODIUM ION | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-04 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)
To Be Published
|
|
8EY4
 
 | | Contact-dependent growth inhibition toxin-immunity protein complex from E. coli O32:H37 | | Descriptor: | Cys_rich_CPCC domain-containing protein, FE (III) ION, PT-VENN domain-containing protein | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Hayes, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Contact-dependent growth inhibition toxin-immunity protein complex from E. coli O32:H37
To Be Published
|
|
8EY3
 
 | | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37 | | Descriptor: | Cys_rich_CPCC domain-containing protein, FE (III) ION, SODIUM ION | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Hayes, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37
To Be Published
|
|
5FFP
 
 | | Crystal structure of CdiI from Burkholderia dolosa AUO158 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Immunity 23 family protein | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2015-12-18 | | Release date: | 2016-01-20 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CdiI from Burkholderia dolosa AUO158
To Be Published
|
|
5DS0
 
 | | Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09 | | Descriptor: | COBALT (II) ION, GLYCEROL, Peptidase M42 | | Authors: | Michalska, K, Chhor, G, Mootz, J, Endres, M, Jedrzejczak, R, Babnigg, G, Steen, A, Lloyd, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09
To Be Published
|
|
6C5C
 
 | | Crystal structure of the 3-dehydroquinate synthase (DHQS) domain of Aro1 from Candida albicans SC5314 in complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate synthase, CHLORIDE ION, ... | | Authors: | Michalska, K, Evdokimova, E, Di Leo, R, Stogios, P.J, Savchenko, A, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-16 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
4TX9
 
 | | Crystal structure of HisAp from Streptomyces sviceus with degraded ProFAR | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Phosphoribosyl isomerase A, SULFATE ION | | Authors: | Michalska, K, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4W9T
 
 | | Crystal structure of HisAP from Streptomyces sp. Mg1 | | Descriptor: | Phosphoribosyl isomerase A, SULFATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
5CQF
 
 | | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae | | Descriptor: | IODIDE ION, L-lysine 6-monooxygenase | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Weerth, R.S, Cao, H, Yennamalli, R, Phillips Jr, G.N, Thomas, M.G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae
To Be Published
|
|
