7T6D
 
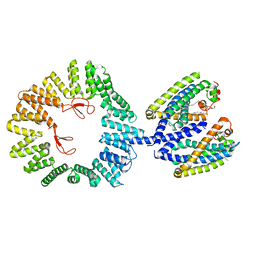 | | CryoEM structure of the YejM/LapB complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Inner membrane protein YejM, ... | | Authors: | Mi, W, Shu, S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Regulatory mechanisms of lipopolysaccharide synthesis in Escherichia coli.
Nat Commun, 13, 2022
|
|
8V24
 
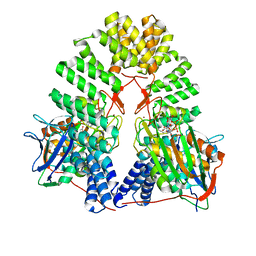 | | LapB cytoplasmic domain in complex with LpxC | | Descriptor: | ACETATE ION, Lipopolysaccharide assembly protein B, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mi, W, Shu, S. | | Deposit date: | 2023-11-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Dual function of LapB (YciM) in regulating Escherichia coli lipopolysaccharide synthesis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5TTP
 
 | |
5V7V
 
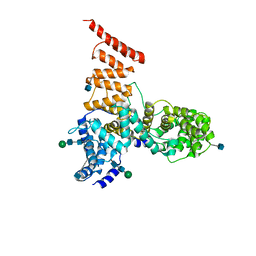 | | Cryo-EM structure of ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ERAD-associated E3 ubiquitin-protein ligase component HRD3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mi, W, Schoebel, S, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
5TV4
 
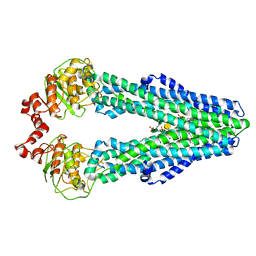 | | 3D cryo-EM reconstruction of nucleotide-free MsbA in lipid nanodisc | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, L-glycero-alpha-D-manno-heptopyranose-(1-7)-L-glycero-alpha-D-manno-heptopyranose-(1-3)-L-glycero-alpha-D-manno-heptopyranose-(1-5)-[3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)]3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-alpha-D-glucopyranose-(1-6)-2-amino-2-deoxy-alpha-D-glucopyranose, LAURIC ACID, ... | | Authors: | Mi, W, Walz, T, Liao, M. | | Deposit date: | 2016-11-08 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of MsbA-mediated lipopolysaccharide transport.
Nature, 549, 2017
|
|
3MXZ
 
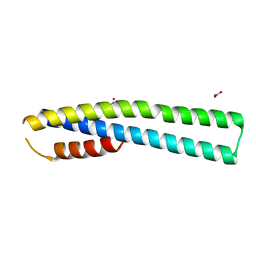 | | Crystal Structure of tubulin folding cofactor A from Arabidopsis thaliana | | Descriptor: | NITRATE ION, Tubulin-specific chaperone A | | Authors: | Lu, L, Nan, J, Mi, W, Su, X.D, Li, Y. | | Deposit date: | 2010-05-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Crystal structure of tubulin folding cofactor A from Arabidopsis thaliana and its beta-tubulin binding characterization
Febs Lett., 584, 2010
|
|
3NR2
 
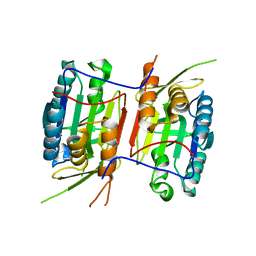 | | Crystal structure of Caspase-6 zymogen | | Descriptor: | Caspase-6 | | Authors: | Su, X.-D, Wang, X.-J, Liu, X, Mi, W, Wang, K.-T. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
2PER
 
 | |
5V6P
 
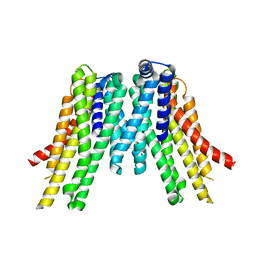 | | CryoEM structure of the ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Authors: | Schoebel, S, Mi, W, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
8DFM
 
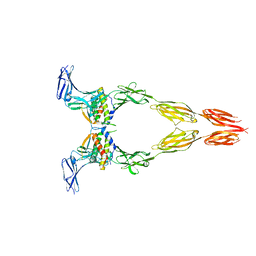 | | Ectodomain of full-length wild-type KIT-SCF dimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Mast/stem cell growth factor receptor Kit, ... | | Authors: | Krimmer, S.G, Bertoletti, N, Mi, W, Schlessinger, J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM analyses of KIT and oncogenic mutants reveal structural oncogenic plasticity and a target for therapeutic intervention.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DFQ
 
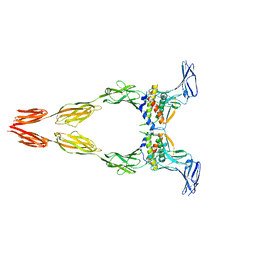 | | Ectodomain of full-length KIT(T417I,delta418-419)-SCF dimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Mast/stem cell growth factor receptor Kit, ... | | Authors: | Krimmer, S.G, Bertoletti, N, Mi, W, Schlessinger, J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM analyses of KIT and oncogenic mutants reveal structural oncogenic plasticity and a target for therapeutic intervention.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DFP
 
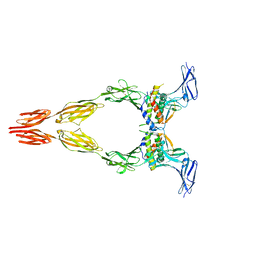 | | Ectodomain of full-length KIT(DupA502,Y503)-SCF dimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Mast/stem cell growth factor receptor Kit, ... | | Authors: | Bertoletti, N, Krimmer, S.G, Mi, W, Schlessinger, J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cryo-EM analyses of KIT and oncogenic mutants reveal structural oncogenic plasticity and a target for therapeutic intervention.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6DBR
 
 | | Cryo-EM structure of RAG in complex with one melted RSS and one unmelted RSS | | Descriptor: | CALCIUM ION, Forward strand of melted RSS substrate DNA, Forward strand of unmelted RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBL
 
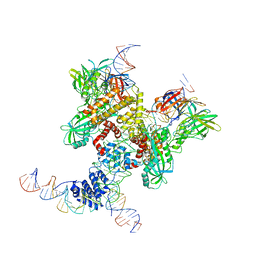 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (5.001 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBQ
 
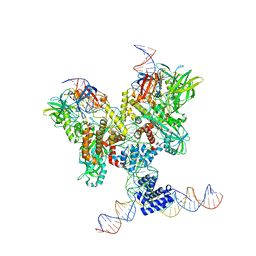 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBW
 
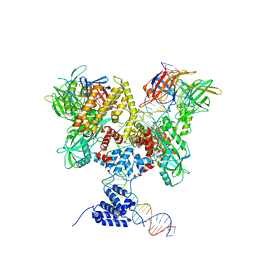 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBT
 
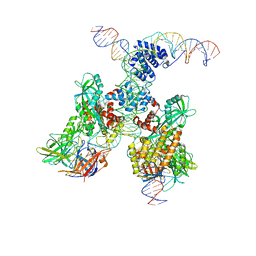 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBX
 
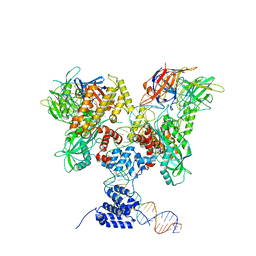 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBV
 
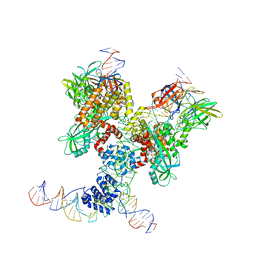 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.291 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBU
 
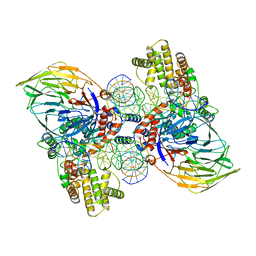 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBI
 
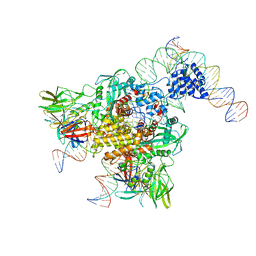 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS signal end, Forward strand of 23-RSS signal end, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBO
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of substrate RSS DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBJ
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward stand of RSS signal end, Forward strand of coding flank, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
