3V11
 
 | |
2QN6
 
 | | Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Translation initiation factor 2 alpha subunit, ... | | Authors: | Mechulam, Y, Yatime, L, Blanquet, S, Schmitt, E. | | Deposit date: | 2007-07-18 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1QQT
 
 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Mechulam, Y, Schmitt, E, Maveyraud, L, Zelwer, C, Nureki, O, Yokoyama, S, Konno, M, Blanquet, S. | | Deposit date: | 1999-06-08 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features.
J.Mol.Biol., 294, 1999
|
|
1KK0
 
 | |
1KK3
 
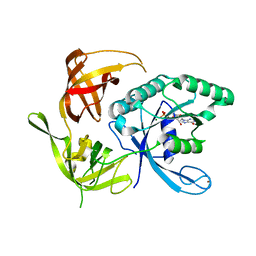 | | Structure of the wild-type large gamma subunit of initiation factor eIF2 from Pyrococcus abyssi complexed with GDP-Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Schmitt, E, Blanquet, S, Mechulam, Y. | | Deposit date: | 2001-12-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors.
EMBO J., 21, 2002
|
|
3M4X
 
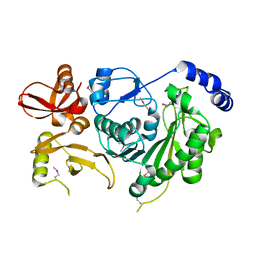 | |
2FMT
 
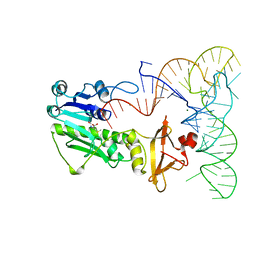 | | METHIONYL-TRNAFMET FORMYLTRANSFERASE COMPLEXED WITH FORMYL-METHIONYL-TRNAFMET | | Descriptor: | FORMYL-METHIONYL-TRNAFMET2, MAGNESIUM ION, METHIONYL-TRNA FMET FORMYLTRANSFERASE, ... | | Authors: | Schmitt, E, Mechulam, Y, Blanquet, S. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet.
EMBO J., 17, 1998
|
|
1XTY
 
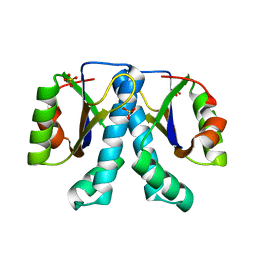 | | Crystal structure of Sulfolobus solfataricus peptidyl-tRNA hydrolase | | Descriptor: | Peptidyl-tRNA hydrolase, SULFATE ION | | Authors: | Fromant, M, Schmitt, E, Mechulam, Y, Lazennec, C, Plateau, P, Blanquet, S. | | Deposit date: | 2004-10-25 | | Release date: | 2005-03-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure at 1.8 A resolution and identification of active site residues of Sulfolobus solfataricus peptidyl-tRNA hydrolase.
Biochemistry, 44, 2005
|
|
2MS9
 
 | | Solution structure of a G-quadruplex | | Descriptor: | DNA (28-MER) | | Authors: | Chung, W.J, Heddi, B, Schmitt, E, Lim, K.W, Mechulam, Y, Phan, A.T. | | Deposit date: | 2014-07-25 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a left-handed DNA G-quadruplex
Proc.Natl.Acad.Sci.USA, 2015
|
|
6Y3G
 
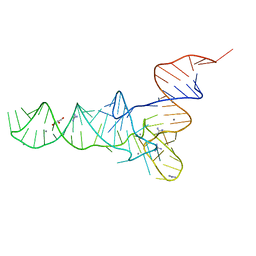 | | Crystal structure of phenylalanine tRNA from Escherichia coli | | Descriptor: | CALCIUM ION, GLYCEROL, GUANIDINE, ... | | Authors: | Bourgeois, G, Mechulam, Y, Schmitt, E. | | Deposit date: | 2020-02-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the interaction between cyclodipeptide synthases and aminoacylated tRNA substrates.
Rna, 26, 2020
|
|
6Q6R
 
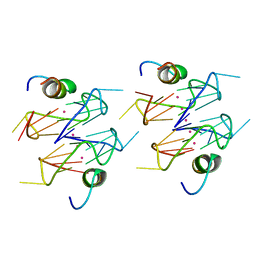 | | Recognition of different base tetrads by RHAU: X-ray crystal structure of G4 recognition motif bound to the 3-end tetrad of a DNA G-quadruplex | | Descriptor: | ATP-dependent DNA/RNA helicase DHX36, POTASSIUM ION, Parallel stranded DNA G-quadruplex | | Authors: | Heddi, B, Cheong, V.V, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2018-12-11 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of different base tetrads by RHAU (DHX36): X-ray crystal structure of the G4 recognition motif bound to the 3'-end tetrad of a DNA G-quadruplex.
J.Struct.Biol., 209, 2020
|
|
6Y4B
 
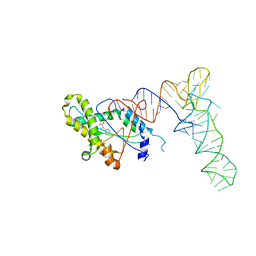 | |
6QJO
 
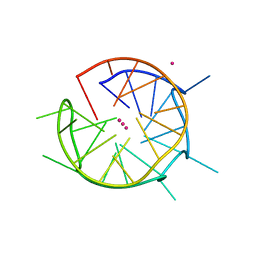 | | DNA containing both right- and left-handed parallel-stranded G-quadruplexes | | Descriptor: | DNA (28-MER), POTASSIUM ION | | Authors: | Winnerdy, F.R, Bakalar, B, Maity, A, Vandana, J.J, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2019-01-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes.
Nucleic Acids Res., 47, 2019
|
|
6JCE
 
 | | NMR solution and X-ray crystal structures of a DNA containing both right-and left-handed parallel-stranded G-quadruplexes | | Descriptor: | 29-mer DNA | | Authors: | Winnerdy, F.R, Bakalar, B, Maity, A, Vandana, J.J, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes.
Nucleic Acids Res., 47, 2019
|
|
6EZ3
 
 | |
6FQ2
 
 | | Structure of minimal sequence for left -handed G-quadruplex formation | | Descriptor: | DNA (5'-D(*GP*TP*GP*GP*TP*GP*GP*TP*GP*GP*TP*G)-3'), POTASSIUM ION | | Authors: | Schmitt, E, Mechulam, Y, Phan, A.T, Heddi, B, Bakalar, B. | | Deposit date: | 2018-02-13 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Minimal Sequence for Left-Handed G-Quadruplex Formation.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6GZ6
 
 | | Structure of a left-handed G-quadruplex | | Descriptor: | DNA (27-MER), POTASSIUM ION | | Authors: | Bakalar, B, Heddi, B, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2018-07-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A Minimal Sequence for Left-Handed G-Quadruplex Formation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
7ZHG
 
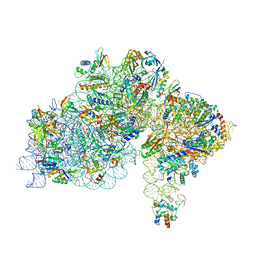 | | High-resolution cryo-EM structure of Pyrococcus abyssi 30S ribosomal subunit bound to mRNA and initiator tRNA anticodon stem-loop | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Kazan, R, Bourgeois, G, Mechulam, Y, Coureux, P.D, Schmitt, E. | | Deposit date: | 2022-04-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
5OCD
 
 | |
5MLP
 
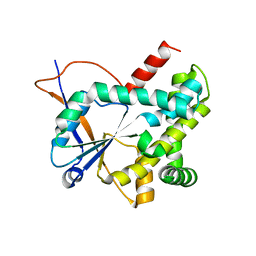 | | Structure of CDPS from Rickettsiella grylli | | Descriptor: | Uncharacterized protein | | Authors: | Bourgeois, G, Seguin, J, Moutiez, M, Babin, M, Belin, P, Mechulam, Y, Gondry, M, Schmitt, E. | | Deposit date: | 2016-12-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for partition of the cyclodipeptide synthases into two subfamilies.
J.Struct.Biol., 203, 2018
|
|
7ZKI
 
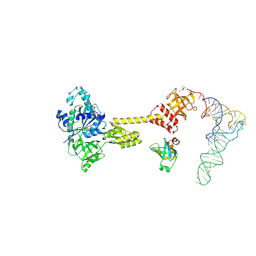 | | Cryo-EM structure of aIF1A:aIF5B:Met-tRNAiMet complex from a Pyrococcus abyssi 30S initiation complex | | Descriptor: | MAGNESIUM ION, METHIONINE, Met-tRNAiMet, ... | | Authors: | Coureux, P.D, Bourgeois, G, Mechulam, Y, Schmitt, E, Kazan, R. | | Deposit date: | 2022-04-13 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
5MLQ
 
 | | Structure of CDPS from Nocardia brasiliensis | | Descriptor: | CDPS, CITRIC ACID | | Authors: | Bourgeois, G, Seguin, J, Moutiez, M, Babin, M, Belin, P, Mechulam, Y, Gondry, M, Schmitt, E. | | Deposit date: | 2016-12-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural basis for partition of the cyclodipeptide synthases into two subfamilies.
J.Struct.Biol., 203, 2018
|
|
8BRW
 
 | | Escherichia coli methionyl-tRNA synthetase mutant L13C,I297C | | Descriptor: | Methionine--tRNA ligase, ZINC ION | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
8BRX
 
 | | Escherichia coli methionyl-tRNA synthetase mutant L13C,I297C complexed with beta-3-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, Methionine--tRNA ligase, ... | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
8BRV
 
 | | Escherichia coli methionyl-tRNA synthetase mutant L13M,I297C complexed with beta3-methionine. | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, Methionine--tRNA ligase, ... | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
