7K3T
 
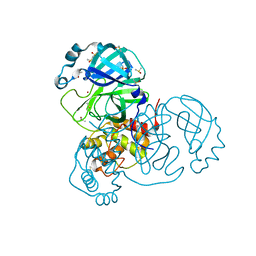 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) at 1.2 A Resolution and a Possible Capture of Zinc Binding Intermediate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-13 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7SEU
 
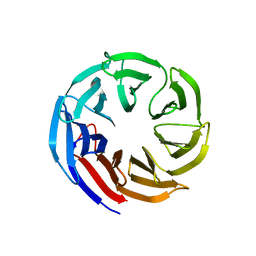 | | Crystal structure of E coli contaminant protein YncE co-purified with a plant protein | | Descriptor: | PQQ-dependent catabolism-associated beta-propeller protein | | Authors: | Chai, L, Zhu, P, Chai, J, Pang, C, Andi, B, McsWeeney, S, Shanklin, J, Liu, Q. | | Deposit date: | 2021-10-01 | | Release date: | 2022-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AlphaFold Protein Structure Database for Sequence-Independent Molecular Replacement
Crystals, 11, 2021
|
|
6TGT
 
 | |
6TB5
 
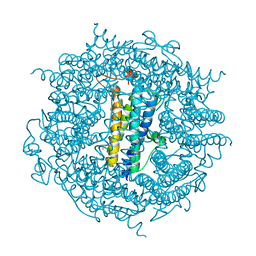 | | The crystal structure of the DPS2 from DEINOCOCCUS RADIODURANS to 1.83A resolution (sequentially soaked in CaCl2 [5mM] for 20 min, then in Ammonium iron(II) sulfate [10mM] for 2h). | | Descriptor: | CALCIUM ION, DNA protection during starvation protein 2, FE (III) ION | | Authors: | Cuypers, M.G, McSweeney, S, Romao, C.V, Mitchell, E.P. | | Deposit date: | 2019-10-31 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of the DPS2 from DEINOCOCCUS RADIODURANS to 1.83A resolution (sequentially soaked in CaCl2 [5mM] for 20 min, then in Ammonium iron(II) sulfate [10mM] for 2h).
To Be Published
|
|
2JH3
 
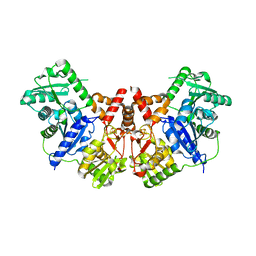 | |
1W3P
 
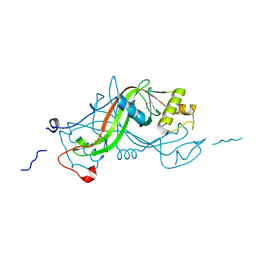 | | NimA from D. radiodurans with a His71-Pyruvate residue | | Descriptor: | ACETATE ION, NIMA-RELATED PROTEIN, PYRUVIC ACID | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W3R
 
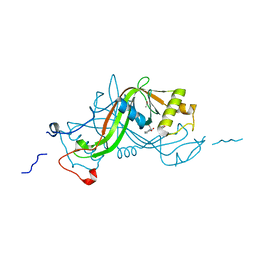 | | NimA from D. radiodurans with Metronidazole and Pyruvate | | Descriptor: | ACETATE ION, Metronidazole, NIMA-RELATED PROTEIN, ... | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W3Q
 
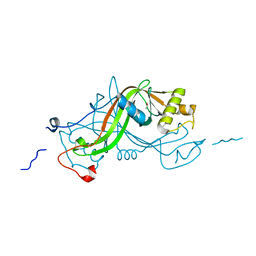 | | NimA from D. radiodurans with covalenly bound lactate | | Descriptor: | ACETATE ION, LACTIC ACID, NIMA-RELATED PROTEIN | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, Mcsweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W3O
 
 | | Crystal structure of NimA from D. radiodurans | | Descriptor: | ACETATE ION, NIMA-RELATED PROTEIN, PYRUVIC ACID | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
7MHF
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHL
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHM
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7K6D
 
 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.48 A Resolution (Cryo-protected) | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K40
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Boceprevir at 1.35 A Resolution | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, boceprevir (bound form) | | Authors: | Kumaran, D, Andi, B, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-14 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K6E
 
 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.63 A Resolution (Direct Vitrification) | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7JYC
 
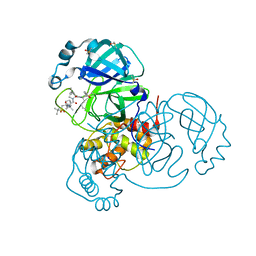 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Narlaprevir | | Descriptor: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-08-30 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
2JGV
 
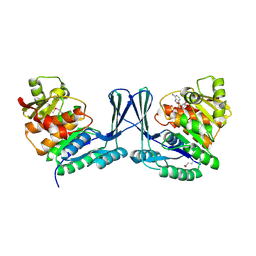 | | STRUCTURE OF Staphylococcus aureus D-TAGATOSE-6-PHOSPHATE KINASE in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TAGATOSE-6-PHOSPHATE KINASE | | Authors: | Miallau, L, Hunter, W.N, McSweeney, S.M, Leonard, G.A. | | Deposit date: | 2007-02-16 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Staphylococcus Aureus D-Tagatose-6-Phosphate Kinase Implicate Domain Motions in Specificity and Mechanism.
J.Biol.Chem., 282, 2007
|
|
1ZIQ
 
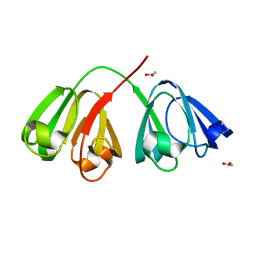 | | Deuterated gammaE crystallin in D2O solvent | | Descriptor: | ACETATE ION, Gamma crystallin E | | Authors: | Artero, J.B, Hartlein, M, McSweeney, S, Timmins, P. | | Deposit date: | 2005-04-27 | | Release date: | 2005-11-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparison of refined X-ray structures of hydrogenated and perdeuterated rat gammaE-crystallin in H2O and D2O.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1HJ9
 
 | | Atomic resolution structures of trypsin provide insight into structural radiation damage | | Descriptor: | ANILINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Leiros, H.-K.S, McSweeney, S.M, Smalas, A.O. | | Deposit date: | 2001-01-10 | | Release date: | 2002-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic Resolution Structure of Trypsin Provide Insight Into Structural Radiation Damage
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HJ8
 
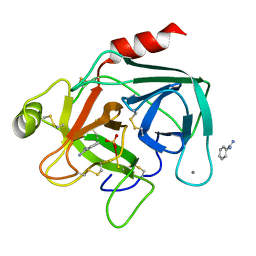 | | 1.00 AA Trypsin from Atlantic Salmon | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Leiros, H.-K.S, Mcsweeney, S.M, Smalas, A.O. | | Deposit date: | 2001-01-09 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic Resolution Structure of Trypsin Provide Insight Into Structural Radiation Damage
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HZZ
 
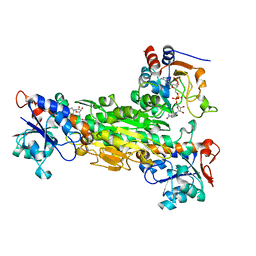 | | THE ASYMMETRIC COMPLEX OF THE TWO NUCLEOTIDE-BINDING COMPONENTS (DI, DIII) OF PROTON-TRANSLOCATING TRANSHYDROGENASE | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTON-TRANSLOCATING NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE SUBUNIT PNTAA, ... | | Authors: | Cotton, N.P.J, White, S.A, Peake, S.J, McSweeney, S, Jackson, J.B. | | Deposit date: | 2001-01-27 | | Release date: | 2001-08-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of an asymmetric complex of the two nucleotide binding components of proton-translocating transhydrogenase.
Structure, 9, 2001
|
|
1I4U
 
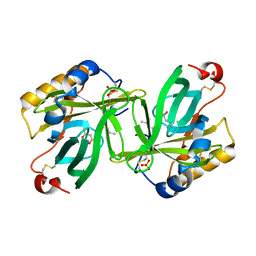 | | THE C1 SUBUNIT OF ALPHA-CRUSTACYANIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CRUSTACYANIN, SULFATE ION | | Authors: | Gordon, E.J, Leonard, G.A, McSweeney, S, Zagalsky, P.F. | | Deposit date: | 2001-02-23 | | Release date: | 2001-09-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The C1 subunit of alpha-crustacyanin: the de novo phasing of the crystal structure of a 40 kDa homodimeric protein using the anomalous scattering from S atoms combined with direct methods.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4C2U
 
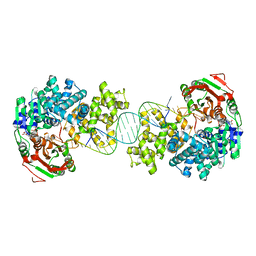 | | Crystal structure of Deinococcus radiodurans UvrD in complex with DNA, Form 1 | | Descriptor: | DNA HELICASE II, FOR25, GLYCEROL, ... | | Authors: | Stelter, M, Acajjaoui, S, McSweeney, S, Timmins, J. | | Deposit date: | 2013-08-20 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Mechanistic Insight Into DNA Unwinding by Deinococcus Radiodurans Uvrd.
Plos One, 8, 2013
|
|
4C30
 
 | | Crystal structure of Deinococcus radiodurans UvrD in complex with DNA, form 2 | | Descriptor: | DNA HELICASE II, DNA STRAND FOR25, DNA STRAND REV25, ... | | Authors: | Stelter, M, Acajjaoui, S, McSweeney, S, Timmins, J. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Mechanistic Insight Into DNA Unwinding by Deinococcus Radiodurans Uvrd.
Plos One, 8, 2013
|
|
