2QCO
 
 | | Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni | | Descriptor: | CmeR, GLYCEROL | | Authors: | Gu, R, Su, C, Shi, F, Li, M, McDermott, G, Zhang, Q, Yu, E.W. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni.
J.Mol.Biol., 372, 2007
|
|
2QOP
 
 | | Crystal structure of the transcriptional regulator AcrR from Escherichia coli | | Descriptor: | HTH-type transcriptional regulator acrR | | Authors: | Li, M, Gu, R, Su, C.-C, McDermott, G, Yu, E.W. | | Deposit date: | 2007-07-20 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the transcriptional regulator AcrR from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
3BCG
 
 | | Conformational changes of the AcrR regulator reveal a mechanism of induction | | Descriptor: | HTH-type transcriptional regulator acrR | | Authors: | Gu, R, Li, M, Su, C.C, Long, F, Yang, F, McDermott, G, Yu, E.Y. | | Deposit date: | 2007-11-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational change of the AcrR regulator reveals a possible mechanism of induction.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1H3D
 
 | | STRUCTURE OF THE E.COLI ATP-PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-PHOSPHORIBOSYLTRANSFERASE, L(+)-TARTARIC ACID | | Authors: | Lohkamp, B, McDermott, G, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-08-27 | | Release date: | 2003-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of Escherichia Coli ATP-Phosphoribosyltransferase: Identification of Substrate Binding Sites and Mode of AMP Inhibition
J.Mol.Biol., 336, 2004
|
|
1KZU
 
 | | INTEGRAL MEMBRANE PERIPHERAL LIGHT HARVESTING COMPLEX FROM RHODOPSEUDOMONAS ACIDOPHILA STRAIN 10050 | | Descriptor: | BACTERIOCHLOROPHYLL A, LIGHT HARVESTING PROTEIN B-800/850, Rhodopin b-D-glucoside | | Authors: | Cogdell, R.J, Freer, A.A, Isaacs, N.W, Hawthornthwaite-Lawless, A.M, Mcdermott, G, Papiz, M.Z, Prince, S.M. | | Deposit date: | 1996-08-31 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Apoprotein structure in the LH2 complex from Rhodopseudomonas acidophila strain 10050: modular assembly and protein pigment interactions.
J.Mol.Biol., 268, 1997
|
|
1KGB
 
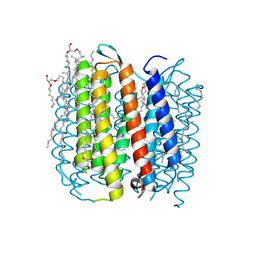 | | structure of ground-state bacteriorhodopsin | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, RETINAL, bacteriorhodopsin | | Authors: | Facciotti, M.T, Rouhani, S, Burkard, F.T, Betancourt, F.M, Downing, K.H, Rose, R.B, McDermott, G, Glaeser, R.M. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle.
Biophys.J., 81, 2001
|
|
1K6W
 
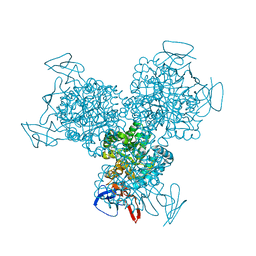 | | The Structure of Escherichia coli Cytosine Deaminase | | Descriptor: | Cytosine Deaminase, FE (III) ION | | Authors: | Ireton, G.C, McDermott, G, Black, M.E, Stoddard, B.L. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of Escherichia coli cytosine deaminase.
J.Mol.Biol., 315, 2002
|
|
1K70
 
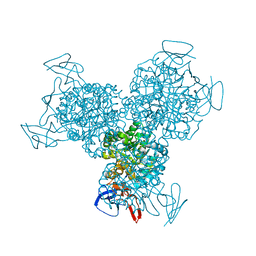 | | The Structure of Escherichia coli Cytosine Deaminase bound to 4-Hydroxy-3,4-Dihydro-1H-Pyrimidin-2-one | | Descriptor: | 4-HYDROXY-3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE, Cytosine Deaminase, FE (III) ION | | Authors: | Ireton, G.C, McDermott, G, Black, M.E, Stoddard, B.L. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Escherichia coli cytosine deaminase.
J.Mol.Biol., 315, 2002
|
|
1KG9
 
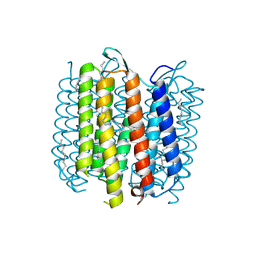 | | Structure of a "mock-trapped" early-M intermediate of bacteriorhosopsin | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, RETINAL, bacteriorhodopsin | | Authors: | Facciotti, M.T, Rouhani, S, Burkard, F.T, Betancourt, F.M, Downing, K.H, Rose, R.B, McDermott, G, Glaeser, R.M. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle.
Biophys.J., 81, 2001
|
|
1KG8
 
 | | X-ray structure of an early-M intermediate of bacteriorhodopsin | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, RETINAL, bacteriorhodopsin | | Authors: | Facciotti, M.T, Rouhani, S, Burkard, F.T, Betancourt, F.M, Downing, K.H, Rose, R.B, McDermott, G, Glaeser, R.M. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle.
Biophys.J., 81, 2001
|
|
1OZN
 
 | | 1.5A Crystal Structure of the Nogo Receptor Ligand Binding Domain Reveals a Convergent Recognition Scaffold Mediating Inhibition of Myelination | | Descriptor: | ACETIC ACID, Reticulon 4 receptor, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, X, Bazan, J.F, Park, J.B, McDermott, G, He, Z, Garcia, K.C. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of the Nogo Receptor Ectodomain. A Recognition module implicated in Myelin Inhibition.
Neuron, 38, 2003
|
|
2HQF
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
2HQD
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
2HQC
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
2HQG
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
1OYE
 
 | | Structural Basis of Multiple Binding Capacity of the AcrB multidrug Efflux Pump | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1OY9
 
 | | Structural Basis of Multiple Drug Binding Capacity of the AcrB Multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B, ETHIDIUM | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1OY8
 
 | | Structural Basis of Multiple Drug Binding Capacity of the AcrB Multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B, RHODAMINE 6G | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1Q1K
 
 | | Structure of ATP-phosphoribosyltransferase from E. coli complexed with PR-ATP | | Descriptor: | ATP phosphoribosyltransferase, L(+)-TARTARIC ACID, PHOSPHORIBOSYL ATP | | Authors: | Lohkamp, B, McDermott, G, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2003-07-21 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of Escherichia coli ATP-phosphoribosyltransferase: identification of substrate binding sites and mode of AMP inhibition
J.Mol.Biol., 336, 2004
|
|
1OY6
 
 | | Structural Basis of the Multiple Binding Capacity of the AcrB Multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1T9U
 
 | |
1T9X
 
 | |
1T9Y
 
 | |
1T9V
 
 | |
1T9W
 
 | | Structural Basis of Multidrug Transport by the AcrB Multidrug Efflux Pump | | Descriptor: | 6-[[(2-ETHOXY-1-NAPHTHALENYL)CARBONYL]AMINO]-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLATE, Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Nikaido, H. | | Deposit date: | 2004-05-18 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | A Periplasmic Drug-Binding Site of the AcrB Multidrug Efflux Pump: a Crystallographic and Site-Directed Mutagenesis Study
J.Bacteriol., 187, 2005
|
|
