4ZDM
 
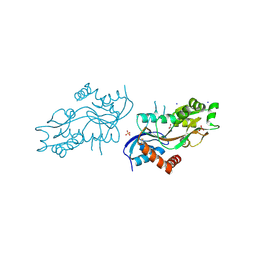 | | Pleurobrachia bachei iGluR3 LBD Glycine Complex | | Descriptor: | GLYCINE, Glutamate receptor kainate-like protein, SODIUM ION, ... | | Authors: | Grey, R.J, Mayer, M.L. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Glycine activated ion channel subunits encoded by ctenophore glutamate receptor genes.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5UN2
 
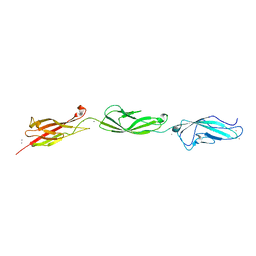 | |
5UZ8
 
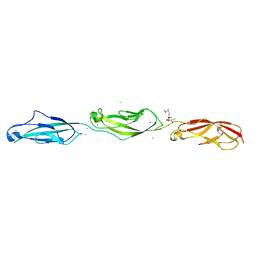 | | Crystal Structure of Mouse Cadherin-23 EC22-24 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cadherin-23, ... | | Authors: | Patel, A, Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
5TPK
 
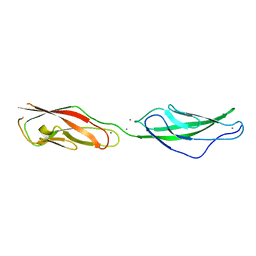 | |
5CYX
 
 | |
5ULU
 
 | |
4KCC
 
 | | Crystal Structure of the NMDA Receptor GluN1 Ligand Binding Domain Apo State | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, PHOSPHATE ION | | Authors: | Berger, A.J, Lau, A.Y, Mayer, M.L. | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Conformational Analysis of NMDA Receptor GluN1, GluN2, and GluN3 Ligand-Binding Domains Reveals Subtype-Specific Characteristics.
Structure, 21, 2013
|
|
3U93
 
 | |
2YMB
 
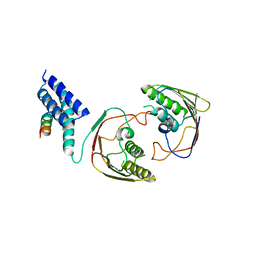 | | Structures of MITD1 | | Descriptor: | CHARGED MULTIVESICULAR BODY PROTEIN 1A, MIT DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Hadders, M.A, Agromayor, M, Obita, T, Perisic, O, Caballe, A, Kloc, M, Lamers, M.H, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5W1D
 
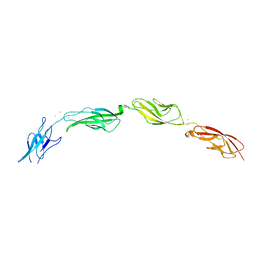 | |
5W4T
 
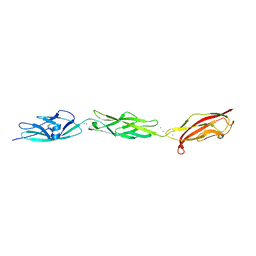 | | Crystal Structure of Fish Cadherin-23 EC1-3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | De-la-Torre, P, Sotomayor, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
4YKP
 
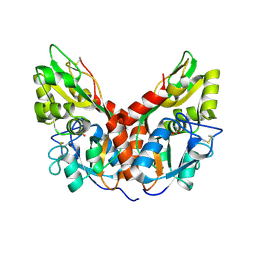 | |
5WJM
 
 | | Crystal Structure of Mouse Cadherin-23 EC17-18 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cadherin-23, ... | | Authors: | De-la-Torre, P, Sotomayor, M. | | Deposit date: | 2017-07-23 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
5ULY
 
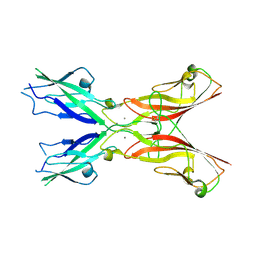 | |
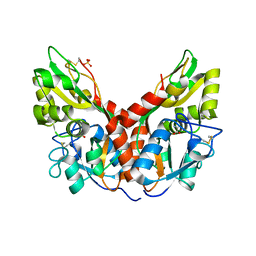
4YKI
 
 | |
5WJ8
 
 | | Crystal Structure of Human Cadherin-23 EC13-14 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cadherin-23, ... | | Authors: | Velez-Cortes, F, Conghui, C, De-la-Torre, P, Sotomayor, M. | | Deposit date: | 2017-07-21 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
4YKJ
 
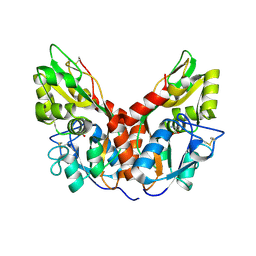 | |
3EBB
 
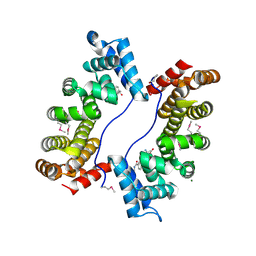 | | PLAP/P97 complex | | Descriptor: | MAGNESIUM ION, PHOSPHOLIPASE A2-ACTIVATING PROTEIN, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE (TER ATP | | Authors: | Walker, J.R, Qiu, L, Akutsu, M, Slessarev, Y, Amaya, M.F, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-27 | | Release date: | 2009-02-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the PLAA/Ufd3-p97/Cdc48 complex.
J.Biol.Chem., 285, 2010
|
|
3U92
 
 | |
3OLZ
 
 | |
3OM0
 
 | |
3OM1
 
 | | Crystal structure of the GluK5 (KA2) ATD dimer at 1.7 Angstrom Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Kumar, J, Mayer, M.L. | | Deposit date: | 2010-08-26 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Crystal Structures of the Glutamate Receptor Ion Channel GluK3 and GluK5 Amino-Terminal Domains.
J.Mol.Biol., 404, 2010
|
|
3QJ6
 
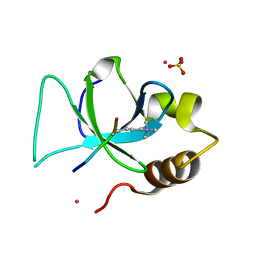 | | The crystal structure of PWWP domain of human Hepatoma-derived growth factor 2 in complex with H3K79me3 peptide | | Descriptor: | H3K79me3, Hepatoma-derived growth factor-related protein 2, SULFATE ION, ... | | Authors: | Zeng, H, Amaya, M.F, Tempel, W, Walker, J.R, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
3QLV
 
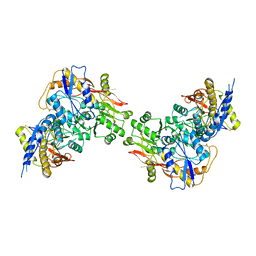 | |
3QKJ
 
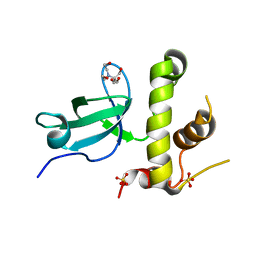 | | The PWWP domain of human DNA (CYTOSINE-5-)-METHYLTRANSFERASE 3 BETA in complex with a bis-tris molecule | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA cytosine-5 methyltransferase 3 beta isoform 6 variant, SULFATE ION | | Authors: | Zeng, H, Amaya, M.F, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
