5EHS
 
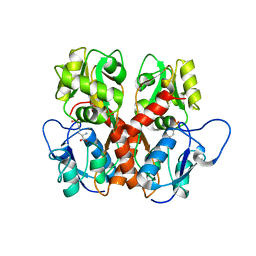 | |
2QS4
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with LY466195 at 1.58 Angstroms resolution | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, AMMONIUM ION, GLYCEROL, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
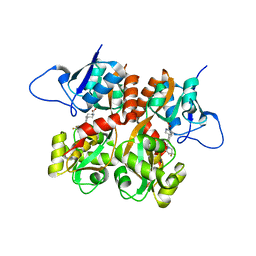
5M3A
 
 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-6-(1-methyl-5-phenoxy-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
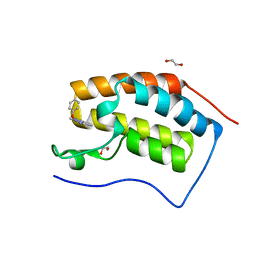
2QS1
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP315 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-4,5-dibromothiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS3
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP316 at 1.76 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-5-phenylthiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | ACET is a highly potent and specific kainate receptor antagonist: characterisation and effects on hippocampal mossy fibre function.
Neuropharmacology, 56, 2009
|
|
2RC7
 
 | |
4XHZ
 
 | |
3U94
 
 | |
5CO1
 
 | |
4IO5
 
 | |
4IO6
 
 | |
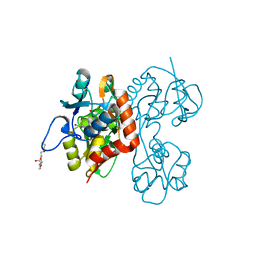
4IO4
 
 | | Crystal Structure of the AvGluR1 ligand binding domain complex with serine at 1.94 Angstrom resolution | | Descriptor: | AvGluR1 ligand binding domain, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lomash, S, Chittori, S, Mayer, M.L. | | Deposit date: | 2013-01-07 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Anions Mediate Ligand Binding in Adineta vaga Glutamate Receptor Ion Channels.
Structure, 21, 2013
|
|
5VH2
 
 | |
4IO7
 
 | |
4YKK
 
 | |
4IO3
 
 | |
4IO2
 
 | |
2RCB
 
 | |
5VVM
 
 | |
5VT8
 
 | |
4KCD
 
 | | Crystal Structure of the NMDA Receptor GluN3A Ligand Binding Domain Apo State | | Descriptor: | GLYCEROL, Glutamate receptor ionotropic, NMDA 3A | | Authors: | Yao, Y, Lau, A.Y, Mayer, M.L. | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conformational Analysis of NMDA Receptor GluN1, GluN2, and GluN3 Ligand-Binding Domains Reveals Subtype-Specific Characteristics.
Structure, 21, 2013
|
|
5ABK
 
 | | Structure of the N-terminal domain of the metalloprotease PrtV from Vibrio cholerae | | Descriptor: | METALLOPROTEASE | | Authors: | Persson, C, Mayzel, M, Edwin, A, Wai, S.N, Ohman, A, Sauer-Eriksson, A.E, Karlsson, G. | | Deposit date: | 2015-08-06 | | Release date: | 2015-08-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-Terminal Domain of the Metalloprotease Prtv from Vibrio Cholerae.
Protein Sci., 24, 2015
|
|
1XK8
 
 | | Divalent cation tolerant protein CUTA from Homo sapiens O60888 | | Descriptor: | Divalent cation tolerant protein CUTA, SODIUM ION | | Authors: | Tempel, W, Chen, L, Liu, Z.-J, Lee, D, Shah, A, Dailey, T.A, Mayer, M.R, Arendall III, W.B, Rose, J.P, Dailey, H.A, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-27 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Divalent cation tolerant protein CUTA from Homo sapiens O60888
To be published
|
|
2RCA
 
 | |
1VKA
 
 | | Southeast Collaboratory for Structural Genomics: Hypothetical Human Protein Q15691 N-Terminal Fragment | | Descriptor: | Microtubule-associated protein RP/EB family member 1 | | Authors: | Liu, Z.-J, Tempel, W, Schubot, F.D, Shah, A, Dailey, T.A, Mayer, M.R, Rose, J.P, Dailey, H.A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-10 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Southeast Collaboratory for Structural Genomics: Hypothetical Human Protein Q15691 N-Terminal Fragment
TO BE PUBLISHED
|
|
