5CYX
 
 | |
4IO5
 
 | |
4IO2
 
 | |
4IO3
 
 | |
6PIM
 
 | | Crystal Structure of Human Protocadherin-1 EC3-4 | | Descriptor: | CALCIUM ION, Protocadherin-1 | | Authors: | Modak, D, Sotomayor, M. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Identification of an adhesive interface for the non-clustered delta 1 protocadherin-1 involved in respiratory diseases.
Commun Biol, 2, 2019
|
|
8V2F
 
 | | Crystal structure of IRAK4 kinase domain with compound 9 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V1O
 
 | | Crystal structure of IRAK4 kinase domain with compound 4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V2L
 
 | | Crystal structure of IRAK4 kinase domain with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-1 receptor-associated kinase 4, N-{2-[4-(hydroxymethyl)phenyl]-6-(2-hydroxypropan-2-yl)-2H-indazol-5-yl}-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
2L3L
 
 | | The solution structure of the N-terminal domain of human Tubulin Binding Cofactor C reveals a platform for the interaction with ab-tubulin | | Descriptor: | Tubulin-specific chaperone C | | Authors: | Garcia-Mayoral, M.F, Castano, R, Lopez-Fanarraga, M.L, Zabala, J.C, Rico, M, Bruix, M. | | Deposit date: | 2010-09-14 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of human tubulin binding cofactor C reveals a platform for tubulin interaction
To be Published
|
|
6NJF
 
 | | Solution NMR Structure of DANCER3-F34A, a rigid and natively folded single mutant of the dynamic protein DANCER-3 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Damry, A.M, Mayer, M.M, Goto, N.K, Chica, R.A. | | Deposit date: | 2019-01-03 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Origin of conformational dynamics in a globular protein.
Commun Biol, 2, 2019
|
|
7N86
 
 | | Crystal Structure of Human Protocadherin-24 EC1-2 Form II | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Modak, D, Gray, M.E, Sotomayor, M. | | Deposit date: | 2021-06-13 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.175 Å) | | Cite: | Heterophilic and homophilic cadherin interactions in intestinal intermicrovillar links are species dependent.
Plos Biol., 19, 2021
|
|
2MAR
 
 | | Solution structure of Ani s 5 Anisakis simplex allergen | | Descriptor: | SXP/RAL-2 family protein | | Authors: | Garcia-Mayoral, M.F, Trevino, M.A, Perez-Pinar, T, Caballero, M.L, Knaute, T, Umpierrez, A, Bruix, M, Rodriguez-Perez, R. | | Deposit date: | 2013-07-17 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Relationships between IgE/IgG4 epitopes, structure and function in Anisakis simplex Ani s 5, a member of the SXP/RAL-2 protein family
Plos Negl Trop Dis, 8, 2014
|
|
8FDW
 
 | | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2, ... | | Authors: | Zhang, J, Shi, W, Cai, Y.F, Zhu, H.S, Peng, H.Q, Voyer, J, Volloch, S.R, Cao, H, Mayer, M.L, Song, K.K, Xu, C, Lu, J.M, Chen, B. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-10 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane.
Nature, 619, 2023
|
|
6Z0P
 
 | | BceF Tyrosine Kinase Domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BceF | | Authors: | Landau, M, Mayer, M, Abd Alhadi, M, Dvir, H. | | Deposit date: | 2020-05-10 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Insights into the Biofilm-Associated BceF Tyrosine Kinase Domain from Burkholderia cepacia .
Biomolecules, 11, 2021
|
|
7SUU
 
 | |
6NW0
 
 | | Crystal Structure Desulfovibrio desulfuricans Nickel-Substituted Rubredoxin | | Descriptor: | NICKEL (II) ION, Rubredoxin | | Authors: | Slater, J.W, Marguet, S.C, Gray, M.E, Sotomayor, M, Shafaat, H.S. | | Deposit date: | 2019-02-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Power of the Secondary Sphere: Modulating Hydrogenase Activity in Nickel-Substituted Rubredoxin
Acs Catalysis, 2019
|
|
6NW1
 
 | | Crystal Structure Desulfovibrio desulfuricans Nickel-Substituted Rubredoxin V37N | | Descriptor: | NICKEL (II) ION, Rubredoxin | | Authors: | Slater, J.W, Marguet, S.C, Gray, M.E, Sotomayor, M, Shafaat, H.S. | | Deposit date: | 2019-02-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Power of the Secondary Sphere: Modulating Hydrogenase Activity in Nickel-Substituted Rubredoxin
Acs Catalysis, 2019
|
|
4WZX
 
 | | ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins | | Descriptor: | COBALT (II) ION, IST1 homolog, SULFATE ION, ... | | Authors: | Caballe, A, Wenzel, D.M, Agromayor, M, Alam, S.L, Skalicky, J.J, Kloc, M, Carlton, J.G, Labrador, L, Sundquist, W.I, Martin-Serrano, J. | | Deposit date: | 2014-11-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3859 Å) | | Cite: | ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins.
Elife, 4, 2015
|
|
4UQ6
 
 | | Electron density map of GluA2em in complex with LY451646 and glutamate | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
1R4Y
 
 | | SOLUTION STRUCTURE OF THE DELETION MUTANT DELTA(7-22) OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | Ribonuclease alpha-sarcin | | Authors: | Garcia-Mayoral, M.F, Garcia-Ortega, L, Lillo, M.P, Santoro, J, Martinez Del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 2003-10-09 | | Release date: | 2004-04-06 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the noncytotoxic {alpha}-sarcin mutant {Delta}(7-22): The importance of the native conformation of peripheral loops for activity.
Protein Sci., 13, 2004
|
|
6BIT
 
 | | SIRPalpha antibody complex | | Descriptor: | KWAR23 Fab heavy chain, KWAR23 Fab light chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Ring, N.G, Herndler-Brandstetter, D, Weiskopf, K, Shan, L, Volkmer, J.P, George, B.M, Lietzenmayer, M, McKenna, K.M, Naik, T.J, McCarty, A, Zheng, Y, Ring, A.M, Flavell, R.A, Weissman, I.L. | | Deposit date: | 2017-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Anti-SIRP alpha antibody immunotherapy enhances neutrophil and macrophage antitumor activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6OAE
 
 | |
8U7I
 
 | | Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex | | Descriptor: | Endonuclease GajA, Gabija Anti-Defense 1, Gabija protein GajB | | Authors: | Antine, S.P, Johnson, A.G, Mooney, S.E, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
7SZ8
 
 | |
5KUF
 
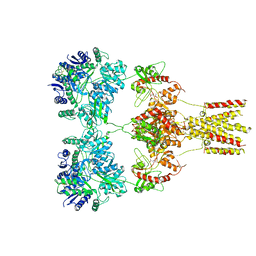 | | GluK2EM with 2S,4R-4-methylglutamate | | Descriptor: | 2S,4R-4-METHYLGLUTAMATE, Glutamate receptor ionotropic, kainate 2 | | Authors: | Meyerson, J.R, Chittori, S, Merk, A, Rao, P, Han, T.H, Serpe, M, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
