1E8H
 
 | |
1E8F
 
 | |
2VAO
 
 | |
6ZLK
 
 | | Equilibrium Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Glucuronic acid/UDP-Galacturonic acid and NAD | | Descriptor: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
9FXQ
 
 | | Ancestral Prenylcysteine Oxidase 1 (PCYOX1) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ancestral Prenylcysteine Oxidase 1 (PCYOX1), FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2024-07-02 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Evolution, structure, and drug-metabolizing activity of mammalian prenylcysteine oxidases.
J.Biol.Chem., 300, 2024
|
|
9FWG
 
 | | LSD1/CoREST bound to bomedemstat | | Descriptor: | Bomedemstat FAD adduct, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Speranzini, V, Mattevi, A. | | Deposit date: | 2024-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of structural, biochemical, pharmacokinetic, and pharmacodynamic properties of the LSD1 inhibitor bomedemstat in preclinical models.
Prostate, 84, 2024
|
|
4UXN
 
 | | LSD1(KDM1A)-CoREST in complex with Z-Pro derivative of MC2580 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, ... | | Authors: | Rodriguez, V, Valente, S, Stazi, G, Lucidi, A, Mercurio, C, Vianello, P, Ciossani, G, Mattevi, A, Botrugno, O.A, Dessanti, P, Minucci, S, Varasi, M, Mai, A. | | Deposit date: | 2014-08-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Pyrrole- and Indole-Containing Tranylcypromine Derivatives as Novel Lysine-Specific Demethylase 1 Inhibitors Active on Cancer Cells
Chemmedchem, 6, 2015
|
|
4UV9
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Ethyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS,10aS)-4a-[(1S,3E)-3-imino-1-phenylpentyl]-7,8-dimethyl-2,4-dioxo-1,3,4,4a,5,10a-hexahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4UVB
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Methyl-Tranylcypromine (1S,2R) | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,7aS)-1-amino-1,10,11-trimethyl-4,6-dioxo-3-phenyl-2,3,5,6,7,7a-hexahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
7QR6
 
 | |
7AL4
 
 | | Ancestral Flavin-containing monooxygenase (FMO) 1 (mammalian) | | Descriptor: | Ancestral Flavin-containing monooxygenase 1 (mammalian), CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nicoll, C.R, Mattevi, A. | | Deposit date: | 2020-10-05 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ancestral reconstruction of mammalian FMO1 enables structural determination, revealing unique features that explain its catalytic properties.
J.Biol.Chem., 296, 2020
|
|
8QNR
 
 | | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: G413N variant in complex with L-gulono-1,4-lactone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-galactono-1,4-lactone dehydrogenase, L-gulono-1,4-lactone | | Authors: | Boverio, A, Mattevi, A. | | Deposit date: | 2023-09-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: G413N variant in complex with L-gulono-1,4-lactone
To Be Published
|
|
8QNB
 
 | | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: in complex with L-galactono-1,4-lactone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-galactono-1,4-lactone, L-galactono-1,4-lactone dehydrogenase | | Authors: | Boverio, A, Mattevi, A. | | Deposit date: | 2023-09-26 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: in complex with L-galactono-1,4-lactone
To Be Published
|
|
8QNC
 
 | |
3PO7
 
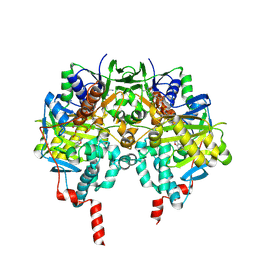 | | Human monoamine oxidase B in complex with zonisamide | | Descriptor: | 1-(1,2-benzoxazol-3-yl)methanesulfonamide, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Aldeco, M, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2010-11-22 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions of monoamine oxidases with the antiepileptic drug zonisamide: specificity of inhibition and structure of the human monoamine oxidase B complex
J.Med.Chem., 54, 2011
|
|
8QMY
 
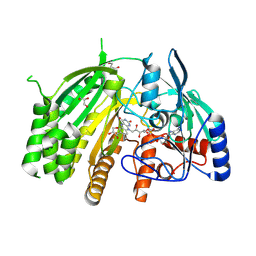 | |
4OVI
 
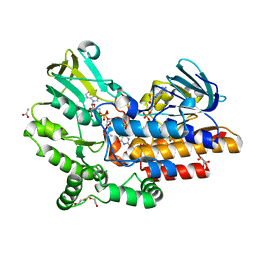 | | Phenylacetone monooxygenase: oxidised enzyme in complex with APADP | | Descriptor: | 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Martinoli, C, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Beyond the Protein Matrix: Probing Cofactor Variants in a Baeyer-Villiger Oxygenation Reaction.
Acs Catalysis, 3, 2013
|
|
8OT8
 
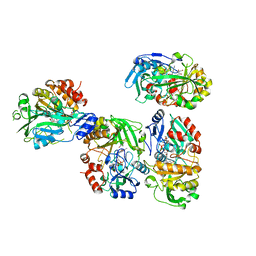 | |
7YWU
 
 | | Eugenol oxidase from rhodococcus jostii: mutant S81H, A423M, H434Y, S394V, I445D, S518P | | Descriptor: | 2-methoxy-4-(prop-2-en-1-yl)phenol, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Alvigini, L, Mattevi, A. | | Deposit date: | 2022-02-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure- and computational-aided engineering of an oxidase to produce isoeugenol from a lignin-derived compound.
Nat Commun, 13, 2022
|
|
7YWV
 
 | | Eugenol oxidase from rhodococcus jostii: mutant S81H, D151E, A423M, H434Y, S394V, Q425S, I445D, S518P | | Descriptor: | 2-methoxy-4-(prop-2-en-1-yl)phenol, CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alvigini, L, Mattevi, A. | | Deposit date: | 2022-02-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure- and computational-aided engineering of an oxidase to produce isoeugenol from a lignin-derived compound.
Nat Commun, 13, 2022
|
|
7ZRY
 
 | |
4F4Q
 
 | | Crystal structure of M. smegmatis DprE1 in complex with FAD and covalently bound BTZ043 | | Descriptor: | 8-(hydroxyamino)-2-[(2S)-2-methyl-1,4-dioxa-8-azaspiro[4.5]dec-8-yl]-6-(trifluoromethyl)-4H-1,3-benzothiazin-4-one, DprE1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Neres, J, Pojer, F, Molteni, E, Chiarelli, L, Riccardi, G, Mattevi, A, Cole, S.T, Binda, C. | | Deposit date: | 2012-05-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Structural Basis for Benzothiazinone-Mediated Killing of Mycobacterium tuberculosis.
Sci Transl Med, 4, 2012
|
|
8Q1G
 
 | | LSD1-CoREST bound to Acetylated K14 of Histone H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 2024
|
|
8Q1H
 
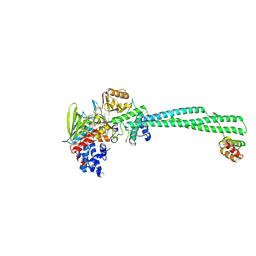 | | LSD1 Y391K-CoREST bound to Histone H3 N-terminal tail | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 2024
|
|
8Q1J
 
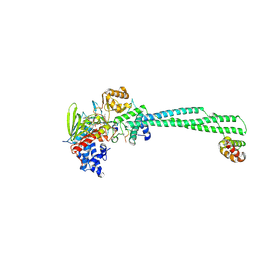 | | LSD1 Y391K-CoREST bound to Acetylated K14 of Histone H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 2024
|
|
