1VBG
 
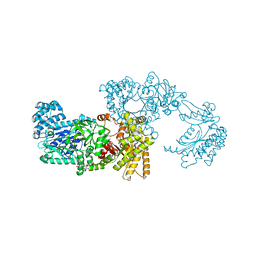 | | Pyruvate Phosphate Dikinase from Maize | | Descriptor: | MAGNESIUM ION, SULFATE ION, pyruvate,orthophosphate dikinase | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
1VBH
 
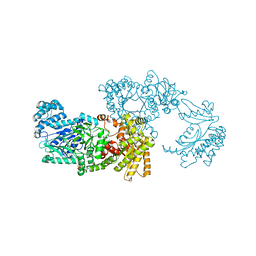 | | Pyruvate Phosphate Dikinase with bound Mg-PEP from Maize | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, SULFATE ION, ... | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
3ED1
 
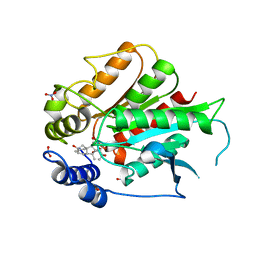 | | Crystal Structure of Rice GID1 complexed with GA3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A3, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-09-02 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
8I09
 
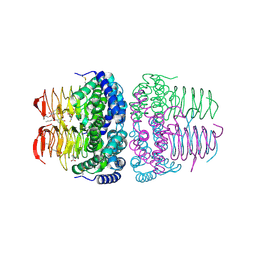 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with butyl gallate | | Descriptor: | CYSTEINE, PHOSPHATE ION, Serine acetyltransferase, ... | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
3EBL
 
 | | Crystal Structure of Rice GID1 complexed with GA4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A4, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-08-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
8I06
 
 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with CoA | | Descriptor: | COENZYME A, CYSTEINE, Serine acetyltransferase | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
8I04
 
 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with serine | | Descriptor: | PHOSPHATE ION, SERINE, Serine acetyltransferase | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
3WZN
 
 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with biotin at 1.3 A resolution | | Descriptor: | BIOTIN, SULFATE ION, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3WZQ
 
 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.7 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, HEXAETHYLENE GLYCOL, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3WZP
 
 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.2 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, GLYCEROL, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3X00
 
 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with bis iminobiotin long tail (Bis-IMNtail) at 1.3 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, ETHANE-1,2-DIAMINE, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design and synthesis of a bivalent iminobiotin analog showing strong affinity toward a low immunogenic streptavidin mutant.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
3WZO
 
 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with biotin long tail (BTNtail) at 1.5 A resolution | | Descriptor: | 6-({5-[(3aS,4S,5S,6aR)-5-oxido-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, CADMIUM ION, GLYCEROL, ... | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
6KU3
 
 | | Crystal structure of gibberellin 2-oxidase3 (GA2ox3)in rice | | Descriptor: | 2-OXOGLUTARIC ACID, GIBBERELLIN A4, GLYCEROL, ... | | Authors: | Takehara, S, Mikami, B, Sakuraba, S, Matsuoka, M, Ueguchi-Tanaka, M. | | Deposit date: | 2019-08-30 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin.
Nat Commun, 11, 2020
|
|
6KUN
 
 | | Crystal structure of dioxygenase for auxin oxidation (DAO) in rice | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent dioxygenase DAO, ... | | Authors: | Takehara, S, Mikami, B, Sakuraba, S, Matsuoka, M, Ueguchi-Tanaka, M. | | Deposit date: | 2019-09-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin.
Nat Commun, 11, 2020
|
|
3AHA
 
 | | Crystal structure of the complex between gp41 fragments N36 and C34 mutant N126K/E137Q | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Transmembrane protein gp41 | | Authors: | Izumi, K, Nakamura, S, Nakano, H, Shimura, K, Sakagami, Y, Oishi, S, Uchiyama, S, Ohkubo, T, Kobayashi, Y, Fujii, N, Matsuoka, M, Kodama, E.N. | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of HIV-1 resistance to a fusion inhibitor, N36, derived from the gp41 amino terminal heptad repeat.
Antiviral Res., 2010
|
|
