8Z9G
 
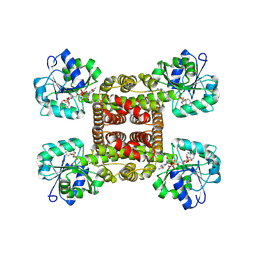 | | Crystal structure of glyoxylate reductase from Acetobacter aceti in complex with NADPH | | Descriptor: | 3-hydroxyisobutyrate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Majumder, T.R, Yoshizawa, T, Inoue, M, Aono, R, Matsumura, H, Mihara, H. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of glyoxylate reductase from Acetobacter aceti
To Be Published
|
|
8Z9F
 
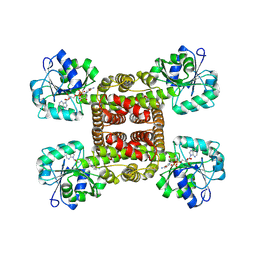 | | Crystal structure of glyoxylate reductase from Acetobacter aceti in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-hydroxyisobutyrate dehydrogenase | | Authors: | Majumder, T.R, Yoshizawa, T, Inoue, M, Aono, R, Matsumura, H, Mihara, H. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of glyoxylate reductase from Acetobacter aceti
To Be Published
|
|
8Z0X
 
 | | Crystal structure of glyoxylate reductase from Acetobacter aceti in the apo form | | Descriptor: | 3-hydroxyisobutyrate dehydrogenase | | Authors: | Majumder, T.R, Yoshizawa, T, Inoue, M, Aono, R, Matsumura, H, Mihara, H. | | Deposit date: | 2024-04-10 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of glyoxylate reductase from Acetobacter aceti
To Be Published
|
|
7XTX
 
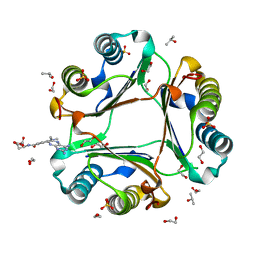 | | High resolution crystal structure of human macrophage migration inhibitory factor in complex with methotrexate | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
7XVX
 
 | |
5E8D
 
 | | Crystal structure of human epiregulin in complex with the Fab fragment of murine monoclonal antibody 9E5 | | Descriptor: | CHLORIDE ION, GLYCEROL, Proepiregulin, ... | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
6L98
 
 | | Crystalline cast nephropathy-causing Bence-Jones protein AK: An entire immunoglobulin lambda light chain dimer | | Descriptor: | Bence-Jones protein lambda light chain AK | | Authors: | Nakagaki, T, Noguchi, K, Yohda, M, Odaka, M, Wakui, H, Matsumura, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Multiple Myeloma-Associated Ig Light Chain Crystalline Cast Nephropathy.
Kidney Int Rep, 5, 2020
|
|
6LL6
 
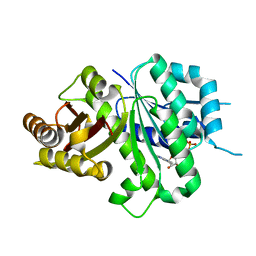 | | Crsyal structure of EcFtsZ (residues 11-316) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yoshizawa, T, Fujita, J, Terakada, H, Ozawa, M, Kuroda, N, Tanaka, S, Uehara, R, Matsumura, H. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the cell-division protein FtsZ from Klebsiella pneumoniae and Escherichia coli.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6LL5
 
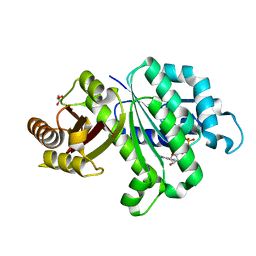 | | Crystal structure of KpFtsZ (residues 11-316) | | Descriptor: | Cell division protein FtsZ, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yoshizawa, T, Fujita, J, Terakado, H, Ozawa, M, Kuroda, N, Tanaka, S, Uehara, R, Matsumura, H. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of the cell-division protein FtsZ from Klebsiella pneumoniae and Escherichia coli.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8KG3
 
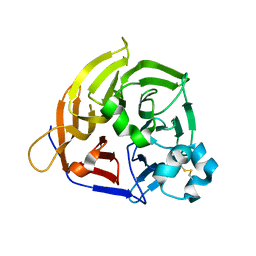 | | Structure of THOUSAND-GRAIN WEIGHT 6 (TGW6) | | Descriptor: | Os06g0623700 protein | | Authors: | Akabane, T, Suzuki, N, Matsumura, H, Yoshizawa, T, Tsuchiya, W, Katoh, E, Hirotsu, N. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | THOUSAND-GRAIN WEIGHT 6, which is an IAA-glucose hydrolase, preferentially recognizes the structure of the indole ring.
Sci Rep, 14, 2024
|
|
7BUX
 
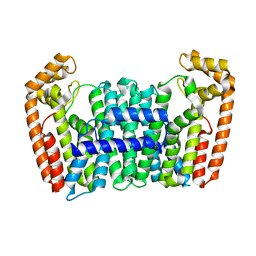 | | Eucommia ulmoides FPS1 | | Descriptor: | FPS2 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
7BUW
 
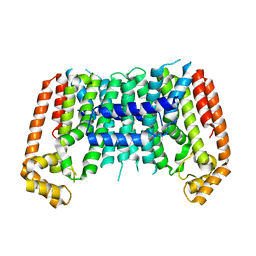 | | Eucommia ulmoides TPT3 mutant -C94Y/A95F | | Descriptor: | FPS3 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
7BUV
 
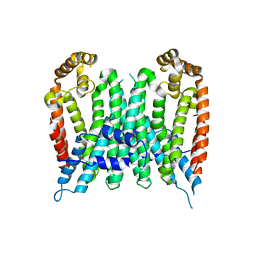 | | Eucommia ulmoides TPT3, crystal form 2 | | Descriptor: | FPS3 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
7BUU
 
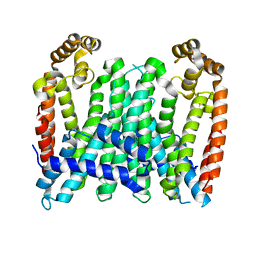 | | Eucommia ulmoides TPT3, crystal form 1 | | Descriptor: | FPS3 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
6M4F
 
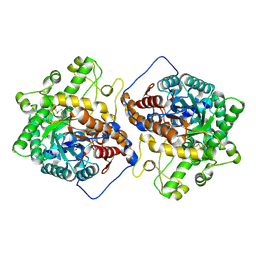 | | Crystal structure of the E496A mutant of HsBglA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase-like enzyme, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
6M4E
 
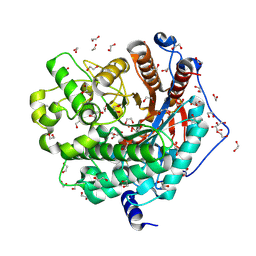 | | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
6M55
 
 | | Crystal structure of the E496A mutant of HsBglA in complex with 4-galactosyllactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase-like enzyme, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-10 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
4O6U
 
 | | 0.89A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
2NVL
 
 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (sulfonic acid form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | Deposit date: | 2006-11-13 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4O6Q
 
 | | 0.95A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (Y75A mutant) | | Descriptor: | FORMIC ACID, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
4O6T
 
 | | 1.25A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant, pH 5.4) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
4O6S
 
 | | 1.32A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant, Zinc Bound) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
8H1O
 
 | | Cryo-EM structure of KpFtsZ-monobody double helical tube | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Mb(Ec/KpFtsZ_S1) | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8IBN
 
 | | Cryo-EM structure of KpFtsZ single filament | | Descriptor: | Cell division protein FtsZ, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, POTASSIUM ION | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hibino, K, Konishi, T, Kato, Y, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
5H5H
 
 | | Staphylococcus aureus FtsZ-GDP R29A mutant in T state | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
