1HUV
 
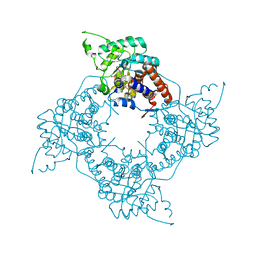 | |
4AAH
 
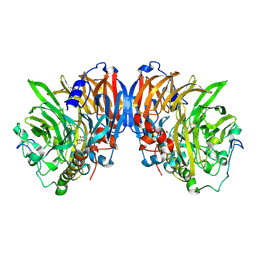 | | METHANOL DEHYDROGENASE FROM METHYLOPHILUS W3A1 | | Descriptor: | CALCIUM ION, METHANOL DEHYDROGENASE, PYRROLOQUINOLINE QUINONE | | Authors: | Mathews, F.S, Xia, Z.-X. | | Deposit date: | 1996-03-10 | | Release date: | 1996-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of the gene sequence and the three-dimensional structure at 2.4 angstroms resolution of methanol dehydrogenase from Methylophilus W3A1.
J.Mol.Biol., 259, 1996
|
|
2TMD
 
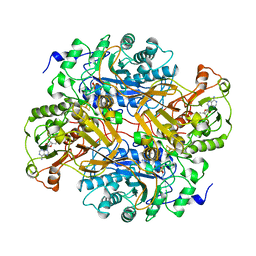 | | CORRELATION OF X-RAY DEDUCED AND EXPERIMENTAL AMINO ACID SEQUENCES OF TRIMETHYLAMINE DEHYDROGENASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Mathews, F.S, Lim, L.W, White, S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Correlation of x-ray deduced and experimental amino acid sequences of trimethylamine dehydrogenase.
J.Biol.Chem., 267, 1992
|
|
3HZL
 
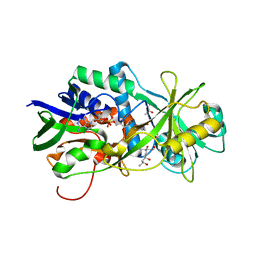 | | Tyr258Phe mutant of NikD, an unusual amino acid oxidase essential for nikkomycin biosynthesis: open form at 1.55A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, NikD protein, ... | | Authors: | Mathews, F.S, Jorns, M.S, Carrell, C.J. | | Deposit date: | 2009-06-23 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Factors that affect oxygen activation and coupling of the two redox cycles in the aromatization reaction catalyzed by NikD, an unusual amino acid oxidase.
Biochemistry, 48, 2009
|
|
1FCB
 
 | |
2PMV
 
 | | Crystal Structure of Human Intrinsic Factor- Cobalamin Complex at 2.6 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COBALAMIN, Gastric intrinsic factor | | Authors: | Mathews, F.S, Gordon, M.M, Chen, Z, Rajashankar, K.R, Ealick, S.E, Alpers, D.H, Sukumar, N. | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human intrinsic factor: Cobalamin complex at 2.6-A resolution
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3M12
 
 | |
3M13
 
 | | Crystal Structure of the Lys265Arg PEG-crystallized mutant of monomeric sarcosine oxidase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase, ... | | Authors: | Mathews, F.S, Chen, Z.-W, Jorns, M.S. | | Deposit date: | 2010-03-04 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of mutations at the oxygen activation site in monomeric sarcosine oxidase.
Biochemistry, 49, 2010
|
|
3M0O
 
 | |
1NWO
 
 | |
1NWP
 
 | |
3SGZ
 
 | | High resolution crystal structure of rat long chain hydroxy acid oxidase in complex with the inhibitor 4-carboxy-5-[(4-chiorophenyl)sulfanyl]-1, 2, 3-thiadiazole. | | Descriptor: | 5-[(4-methylphenyl)sulfanyl]-1,2,3-thiadiazole-4-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 2 | | Authors: | Chen, Z, Vignaud, C, Jaafar, A, Gueritte, F, Guenard, D, Lederer, F, Mathews, F.S. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution crystal structure of rat long chain hydroxy acid oxidase in complex with the inhibitor 4-carboxy-5-[(4-chlorophenyl)sulfanyl]-1, 2, 3-thiadiazole. Implications for inhibitor specificity and drug design.
Biochimie, 94, 2012
|
|
2BBK
 
 | |
2PUX
 
 | | Crystal structure of murine thrombin in complex with the extracellular fragment of murine PAR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 3, Thrombin heavy chain, ... | | Authors: | Bah, A, Chen, Z, Bush-Pelc, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2A7P
 
 | | Crystal Structure of the G81A mutant of the Active Chimera of (S)-Mandelate Dehydrogenase in complex with its substrate 3-indolelactate | | Descriptor: | (S)-Mandelate Dehydrogenase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(INDOL-3-YL) LACTATE, ... | | Authors: | Sukumar, N, Xu, Y, Mitra, B, Mathews, F.S. | | Deposit date: | 2005-07-05 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the G81A mutant form of the active chimera of (S)-mandelate dehydrogenase and its complex with two of its substrates.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3BEF
 
 | | Crystal structure of thrombin bound to the extracellular fragment of PAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 1, Prothrombin | | Authors: | Gandhi, P.S, Bah, A, Chen, Z, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-11-17 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural identification of the pathway of long-range communication in an allosteric enzyme.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2RAC
 
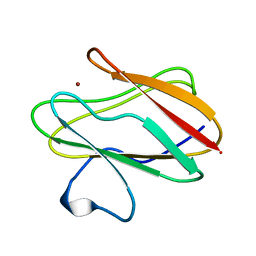 | | AMICYANIN REDUCED, PH 7.7, 1.3 ANGSTROMS | | Descriptor: | COPPER (I) ION, PROTEIN (AMICYANIN) | | Authors: | Cunane, L.M, Chen, Z.-W, Durley, R.C.E, Mathews, F.S. | | Deposit date: | 1998-10-02 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for interprotein complex-dependent effects on the redox properties of amicyanin.
Biochemistry, 37, 1998
|
|
2QDV
 
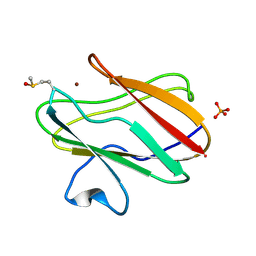 | | Structure of the Cu(II) form of the M51A mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION | | Authors: | Carrell, C.J, Ma, J.K, Wang, Y, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2007-06-21 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | A single methionine residue dictates the kinetic mechanism of interprotein electron transfer from methylamine dehydrogenase to amicyanin.
Biochemistry, 46, 2007
|
|
256B
 
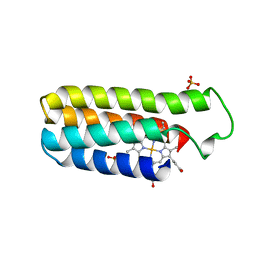 | |
2QDW
 
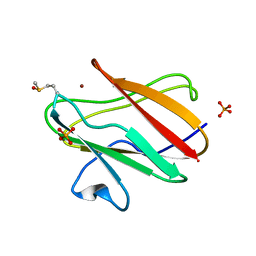 | | Structure of Cu(I) form of the M51A mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (I) ION, PHOSPHATE ION | | Authors: | Ma, J.K, Wang, Y, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2007-06-21 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | A single methionine residue dictates the kinetic mechanism of interprotein electron transfer from methylamine dehydrogenase to amicyanin.
Biochemistry, 46, 2007
|
|
3BEI
 
 | | Crystal structure of the slow form of thrombin in a self_inhibited conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Prothrombin | | Authors: | Gandhi, P.S, Chen, Z, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-11-19 | | Release date: | 2007-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural identification of the pathway of long-range communication in an allosteric enzyme.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BHF
 
 | | Crystal structure of R49K mutant of Monomeric Sarcosine Oxidase crystallized in PEG as precipitant | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase | | Authors: | Hassan-Abdallah, A, Zhao, G, Chen, Z, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2007-11-28 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Arginine 49 is a bifunctional residue important in catalysis and biosynthesis of monomeric sarcosine oxidase: a context-sensitive model for the electrostatic impact of arginine to lysine mutations.
Biochemistry, 47, 2008
|
|
3BHK
 
 | | Crystal structure of R49K mutant of monomeric sarcosine oxidase crystallized in phosphate as precipitant | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Hassan-Abdallah, A, Zhao, G, Chen, Z, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2007-11-28 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Arginine 49 is a bifunctional residue important in catalysis and biosynthesis of monomeric sarcosine oxidase: a context-sensitive model for the electrostatic impact of arginine to lysine mutations.
Biochemistry, 47, 2008
|
|
2Q6U
 
 | | SeMet-substituted form of NikD | | Descriptor: | BENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NikD protein | | Authors: | Carrell, C.J, Bruckner, R.C, Venci, D, Zhao, G, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2007-06-05 | | Release date: | 2007-07-31 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NikD, an Unusual Amino Acid Oxidase Essential for Nikkomycin Biosynthesis: Structures of Closed and Open Forms at 1.15 and 1.90 A Resolution
Structure, 15, 2007
|
|
2MTA
 
 | | CRYSTAL STRUCTURE OF A TERNARY ELECTRON TRANSFER COMPLEX BETWEEN METHYLAMINE DEHYDROGENASE, AMICYANIN AND A C-TYPE CYTOCHROME | | Descriptor: | AMICYANIN, COPPER (II) ION, CYTOCHROME C551I, ... | | Authors: | Chen, L, Mathews, F.S. | | Deposit date: | 1993-10-26 | | Release date: | 1994-01-31 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an electron transfer complex: methylamine dehydrogenase, amicyanin, and cytochrome c551i.
Science, 264, 1994
|
|
