7D5R
 
 | | Structure of the Ca2+-bound C646A mutant of peptidylarginine deiminase type III (PAD3) | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mashimo, R, Akimoto, M, Unno, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
5HP5
 
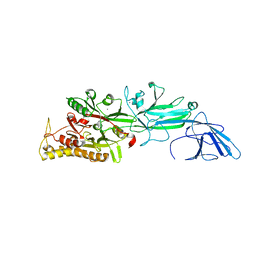 | | Srtucture of human peptidylarginine deiminase type I (PAD1) | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-1 | | Authors: | Unno, M, Nagai, A, Saijo, S, Shimizu, N, Kinjo, S, Mashimo, R, Kizawa, K, Takahara, H. | | Deposit date: | 2016-01-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Monomeric Form of Peptidylarginine Deiminase Type I Revealed by X-ray Crystallography and Small-Angle X-ray Scattering
J.Mol.Biol., 428, 2016
|
|
7D5V
 
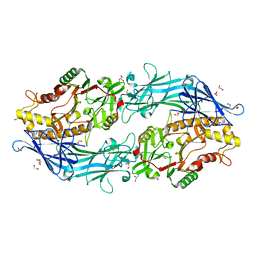 | | Structure of the C646A mutant of peptidylarginine deiminase type III (PAD3) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein-arginine deiminase type-3 | | Authors: | Akimoto, M, Mashimo, R, Unno, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
