2Y2J
 
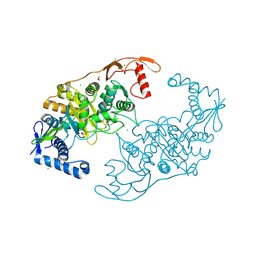 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (ZA4) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | 著者 | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | 登録日 | 2010-12-15 | | 公開日 | 2011-08-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol, 6, 2011
|
|
2Y2K
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (ZA5) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | 著者 | Contreras-Martel, C, Amoroso, A, Woon, E, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C, Dessen, A. | | 登録日 | 2010-12-15 | | 公開日 | 2011-08-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol, 6, 2011
|
|
2Y2G
 
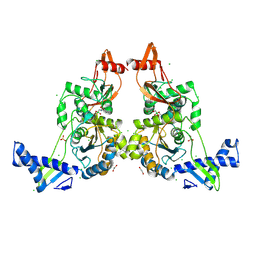 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (A01) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | 著者 | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | 登録日 | 2010-12-15 | | 公開日 | 2011-08-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2N
 
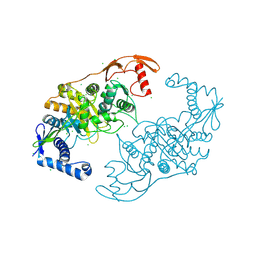 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (E07) | | 分子名称: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | 著者 | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | 登録日 | 2010-12-15 | | 公開日 | 2011-08-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
1VS0
 
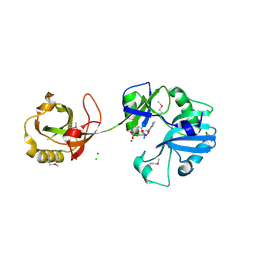 | | Crystal Structure of the Ligase Domain from M. tuberculosis LigD at 2.4A | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Putative DNA ligase-like protein Rv0938/MT0965, ... | | 著者 | Akey, D, Martins, A, Aniukwu, J, Glickman, M.S, Shuman, S, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2006-01-27 | | 公開日 | 2006-02-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure and Nonhomologous End-joining Function of the Ligase Component of Mycobacterium DNA Ligase D.
J.Biol.Chem., 281, 2006
|
|
5W83
 
 | | Rpn8/Rpn11 dimer complex | | 分子名称: | 26S proteasome regulatory subunit RPN8, Ubiquitin carboxyl-terminal hydrolase RPN11, ZINC ION | | 著者 | Dong, K.C, Worden, E.J, Martin, A. | | 登録日 | 2017-06-21 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.554 Å) | | 主引用文献 | An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome.
Mol. Cell, 67, 2017
|
|
6XF9
 
 | | Crystal structure of KSHV ORF68 | | 分子名称: | Packaging protein UL32, ZINC ION | | 著者 | Didychuk, A.L, Gates, S.N, Martin, A, Glaunsinger, B. | | 登録日 | 2020-06-15 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | A pentameric protein ring with novel architecture is required for herpesviral packaging.
Elife, 10, 2021
|
|
6XFA
 
 | | Cryo-EM structure of EBV BFLF1 | | 分子名称: | Packaging protein UL32, ZINC ION | | 著者 | Didychuk, A.L, Gates, S.N, Martin, A, Glaunsinger, B. | | 登録日 | 2020-06-15 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | A pentameric protein ring with novel architecture is required for herpesviral packaging.
Elife, 10, 2021
|
|
4ETC
 
 | | Lysozyme, room temperature, 24 kGy dose | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.906 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETB
 
 | | lysozyme, room temperature, 200 kGy dose | | 分子名称: | CHLORIDE ION, Lysozyme C | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.908 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETE
 
 | | Lysozyme, room-temperature, rotating anode, 0.0021 MGy | | 分子名称: | CHLORIDE ION, Lysozyme C | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.905 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETA
 
 | | Lysozyme, room temperature, 400 kGy dose | | 分子名称: | CHLORIDE ION, Lysozyme C | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ET9
 
 | | Hen egg-white lysozyme solved from 5 fs free-electron laser pulse data | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETD
 
 | | Lysozyme, room-temperature, rotating anode, 0.0026 MGy | | 分子名称: | CHLORIDE ION, Lysozyme C | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.904 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ET8
 
 | | Hen egg-white lysozyme solved from 40 fs free-electron laser pulse data | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
6EF0
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (1D* motor state) | | 分子名称: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | 著者 | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | 登録日 | 2018-08-15 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.43 Å) | | 主引用文献 | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
6EF2
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (5T motor state) | | 分子名称: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | 著者 | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | 登録日 | 2018-08-15 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.27 Å) | | 主引用文献 | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
6EF3
 
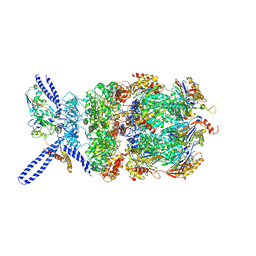 | | Yeast 26S proteasome bound to ubiquitinated substrate (4D motor state) | | 分子名称: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | 著者 | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | 登録日 | 2018-08-15 | | 公開日 | 2018-10-17 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
6EF1
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (5D motor state) | | 分子名称: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | 著者 | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | 登録日 | 2018-08-15 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.73 Å) | | 主引用文献 | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
3J47
 
 | | Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid | | 分子名称: | 26S proteasome regulatory subunit RPN11, 26S proteasome regulatory subunit RPN12, 26S proteasome regulatory subunit RPN3, ... | | 著者 | Estrin, E, Lopez-Blanco, J.R, Chacon, P, Martin, A. | | 登録日 | 2013-06-27 | | 公開日 | 2013-08-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | Formation of an Intricate Helical Bundle Dictates the Assembly of the 26S Proteasome Lid.
Structure, 21, 2013
|
|
3JCK
 
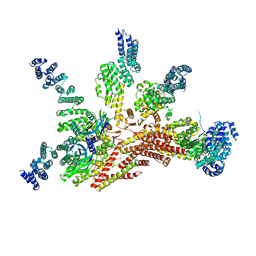 | | Structure of the yeast 26S proteasome lid sub-complex | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN12, 26S proteasome regulatory subunit RPN3, ... | | 著者 | Herzik Jr, M.A, Dambacher, C.M, Worden, E.J, Martin, A, Lander, G.C. | | 登録日 | 2015-12-20 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition.
Elife, 5, 2016
|
|
3HWS
 
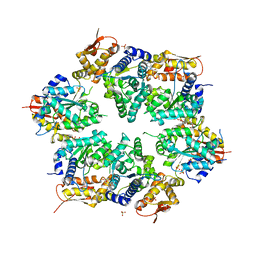 | | Crystal structure of nucleotide-bound hexameric ClpX | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit clpX, MAGNESIUM ION, ... | | 著者 | Glynn, S.E, Martin, A, Baker, T.A, Sauer, R.T. | | 登録日 | 2009-06-18 | | 公開日 | 2009-11-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine.
Cell(Cambridge,Mass.), 139, 2009
|
|
3HTE
 
 | | Crystal structure of nucleotide-free hexameric ClpX | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit clpX, SULFATE ION | | 著者 | Glynn, S.E, Martin, A, Baker, T.A, Sauer, R.T. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (4.026 Å) | | 主引用文献 | Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine.
Cell(Cambridge,Mass.), 139, 2009
|
|
7Q48
 
 | |
7Q6L
 
 | |
