2Y2L
 
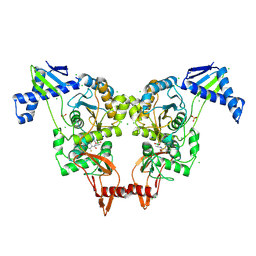 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (E06) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2P
 
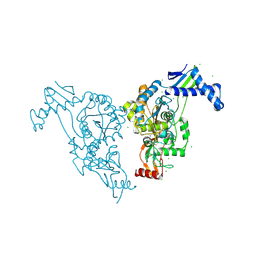 | | Penicillin-binding protein 1b (pbp-1b) in complex with an alkyl boronate (z10) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2M
 
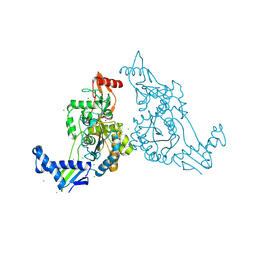 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (E08) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
1YN9
 
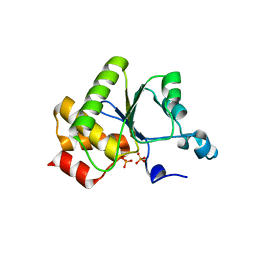 | | Crystal structure of baculovirus RNA 5'-phosphatase complexed with phosphate | | Descriptor: | PHOSPHATE ION, polynucleotide 5'-phosphatase | | Authors: | Changela, A, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of baculovirus RNA triphosphatase complexed with phosphate
J.Biol.Chem., 280, 2005
|
|
5DTL
 
 | |
5DTX
 
 | | Crystal structure of rsEGFP2 in the fluorescent on-state | | Descriptor: | Green fluorescent protein | | Authors: | Adam, V, Martins, A. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rational design of ultrastable and reversibly photoswitchable fluorescent proteins for super-resolution imaging of the bacterial periplasm.
Sci Rep, 6, 2016
|
|
6L8U
 
 | | Crystal structure of human BCDIN3D in complex with SAH | | Descriptor: | RNA 5'-monophosphate methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, Y, Martinez, A, Yamashita, S, Tomita, K. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.925 Å) | | Cite: | Crystal structure of human cytoplasmic tRNAHis-specific 5'-monomethylphosphate capping enzyme.
Nucleic Acids Res., 48, 2020
|
|
1VS0
 
 | | Crystal Structure of the Ligase Domain from M. tuberculosis LigD at 2.4A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative DNA ligase-like protein Rv0938/MT0965, ... | | Authors: | Akey, D, Martins, A, Aniukwu, J, Glickman, M.S, Shuman, S, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-01-27 | | Release date: | 2006-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Nonhomologous End-joining Function of the Ligase Component of Mycobacterium DNA Ligase D.
J.Biol.Chem., 281, 2006
|
|
5OC1
 
 | | Crystal structure of aryl-alcohol oxidase from Pleurotus eryngii in complex with p-anisic acid | | Descriptor: | 4-METHOXYBENZOIC ACID, Aryl-alcohol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Carro, J, Martinez-Julvez, M, Medina, M, Martinez, A, Ferreira, P. | | Deposit date: | 2017-06-29 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein dynamics promote hydride tunnelling in substrate oxidation by aryl-alcohol oxidase.
Phys Chem Chem Phys, 19, 2017
|
|
7ZUI
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 5Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2R)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUH
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) Streptococcus pneumoniae R6 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Penicillin-binding protein 1b | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUJ
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 6Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2S)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUL
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with 8Az lactone - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2R)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-3-phenyl-propan-2-yl]hexanamide, CHLORIDE ION, Penicillin-binding protein 1b | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUK
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 7Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2S)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-3-phenyl-propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7OPT
 
 | | Crystal structure of Trypanosoma cruzi peroxidase | | Descriptor: | Ascorbate peroxidase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Freeman, S.L, Kwon, H, Skafar, V, Fielding, A.J, Martinez, A, Piacenza, L, Radi, R, Raven, E.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of Trypanosoma cruzi heme peroxidase and characterization of its substrate specificity and compound I intermediate.
J.Biol.Chem., 298, 2022
|
|
7OQR
 
 | | Crystal structure of Trypanosoma cruzi peroxidase | | Descriptor: | ACETATE ION, Ascorbate peroxidase, GLYCEROL, ... | | Authors: | Freeman, S.L, Kwon, H, Skafar, V, Fielding, A.J, Martinez, A, Piacenza, L, Radi, R, Raven, E.L. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of Trypanosoma cruzi heme peroxidase and characterization of its substrate specificity and compound I intermediate.
J.Biol.Chem., 298, 2022
|
|
7PIM
 
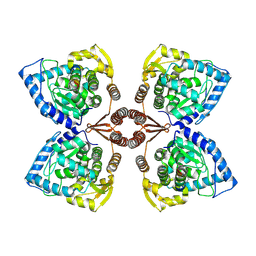 | | Partial structure of tyrosine hydroxylase lacking the first 35 residues in complex with dopamine. | | Descriptor: | FE (III) ION, L-DOPAMINE, Regulatory domain alpha-helix, ... | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7KM3
 
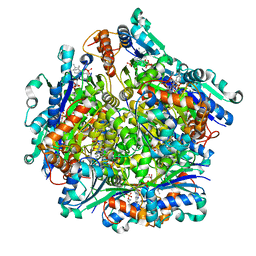 | | Dodecameric Structure of the Chlamydia trachomatis Flavin Prenyltransferase UbiX Ortholog CT220 with FMN and DMAP | | Descriptor: | Dimethylallyl monophosphate, FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX | | Authors: | Nguyen, T, Nicely, N.I, Belaia-Martiniouk, A, Dunne, A.P, McCafferty, D.G. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Functional and structural validation of CT220 as the UbiX-like flavin prenyltransferase from Chlamydial menaquinone biosynthesis
To be published
|
|
7KM2
 
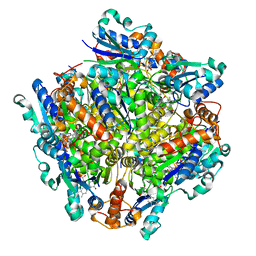 | | Dodecameric Structure of the Chlamydia trachomatis Flavin Prenyltransferase UbiX Ortholog CT220 with FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX | | Authors: | Nguyen, T, Nicely, N.I, Belaia-Martiniouk, A, Dunne, A.P, McCafferty, D.G. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Functional and structural validation of CT220 as the UbiX-like flavin prenyltransferase from Chlamydial menaquinone biosynthesis
To be published
|
|
8PND
 
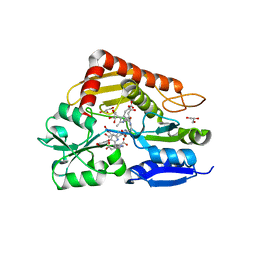 | | The ES3 intermediate of hydroxymethylbilane synthase R167Q variant | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, ... | | Authors: | Saeter, M.C, Bustad, H.J, Laitaoja, M, Janis, J, Martinez, A, Aarsand, A.K, Kallio, J.P. | | Deposit date: | 2023-06-30 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | One ring closer to a closure: the crystal structure of the ES 3 hydroxymethylbilane synthase intermediate.
Febs J., 291, 2024
|
|
7AAJ
 
 | | Human porphobilinogen deaminase in complex with cofactor | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, GLYCEROL, Porphobilinogen deaminase | | Authors: | Kallio, J.P, Bustad, H.J, Martinez, A. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of porphobilinogen deaminase mutants reveals that arginine-173 is crucial for polypyrrole elongation mechanism.
Iscience, 24, 2021
|
|
7AAK
 
 | | Human porphobilinogen deaminase R173W mutant crystallized in the ES2 intermediate state | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, Porphobilinogen deaminase | | Authors: | Kallio, J.P, Bustad, H.J, Martinez, A. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of porphobilinogen deaminase mutants reveals that arginine-173 is crucial for polypyrrole elongation mechanism.
Iscience, 24, 2021
|
|
4J6S
 
 | |
8BA0
 
 | | Drosophila melanogaster complex I in the Twisted state (Dm2) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Agip, A.N.A, Chung, I, Sanchez-Martinez, A, Whitworth, A.J, Hirst, J. | | Deposit date: | 2022-10-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM structures of mitochondrial respiratory complex I from Drosophila melanogaster.
Elife, 12, 2023
|
|
8B9Z
 
 | | Drosophila melanogaster complex I in the Active state (Dm1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Agip, A.A, Chung, I, Sanchez-Martinez, A, Whitworth, A.J, Hirst, J. | | Deposit date: | 2022-10-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures of mitochondrial respiratory complex I from Drosophila melanogaster.
Elife, 12, 2023
|
|
