1SZC
 
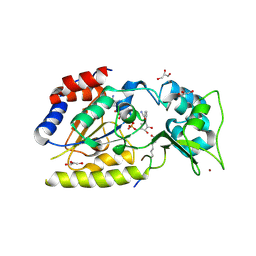 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2RC4
 
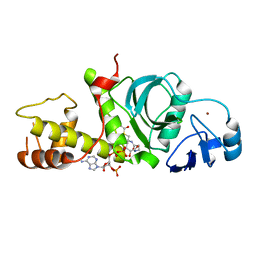 | | Crystal Structure of the HAT domain of the human MOZ protein | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase MYST3, ZINC ION | | Authors: | Holbert, M.A, Sikorski, T, Snowflack, D, Marmorstein, R. | | Deposit date: | 2007-09-19 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human monocytic leukemia zinc finger histone acetyltransferase domain contains DNA-binding activity implicated in chromatin targeting.
J.Biol.Chem., 282, 2007
|
|
8G0L
 
 | |
1QP9
 
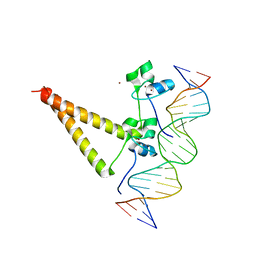 | | STRUCTURE OF HAP1-PC7 COMPLEXED TO THE UAS OF CYC7 | | Descriptor: | CYP1(HAP1-PC7) ACTIVATORY PROTEIN, DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), ... | | Authors: | Lukens, A, King, D, Marmorstein, R. | | Deposit date: | 1999-06-01 | | Release date: | 2000-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HAP1-PC7 bound to DNA: implications for DNA recognition and allosteric effects of DNA-binding on transcriptional activation.
Nucleic Acids Res., 28, 2000
|
|
2B9D
 
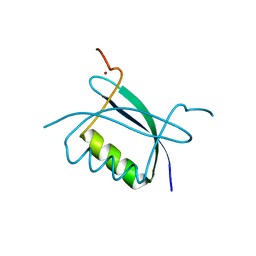 | | Crystal Structure of HPV E7 CR3 domain | | Descriptor: | E7 protein, ZINC ION | | Authors: | Liu, X, Clements, A, Zhao, K, Marmorstein, R. | | Deposit date: | 2005-10-11 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor.
J.Biol.Chem., 281, 2006
|
|
2R7G
 
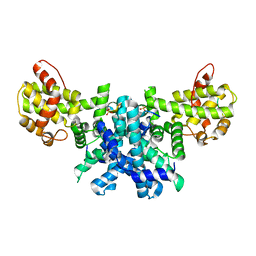 | |
4E26
 
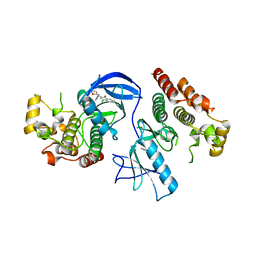 | | BRAF in complex with an organic inhibitor 7898734 | | Descriptor: | 5-chloro-7-[(R)-furan-2-yl(pyridin-2-ylamino)methyl]quinolin-8-ol, Serine/threonine-protein kinase B-raf | | Authors: | Qin, J, Xie, P, Ventocilla, C, Zhou, G, Vultur, A, Chen, Q, Herlyn, M, Winkler, J, Marmorstein, R. | | Deposit date: | 2012-03-07 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of a Novel Family of BRAF(V600E) Inhibitors.
J.Med.Chem., 55, 2012
|
|
5WJD
 
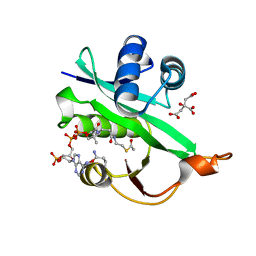 | | Crystal structure of Naa80 bound to acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, CG8481, isoform B, ... | | Authors: | Goris, M, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2017-07-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5WJE
 
 | | Crystal structure of Naa80 bound to a bisubstrate analogue | | Descriptor: | Actin N-terminus peptide, CARBOXYMETHYL COENZYME *A, CG8481, ... | | Authors: | Goris, M, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2017-07-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4KVO
 
 | |
4KVM
 
 | |
5WI2
 
 | | Crystal structure of the KA1 domain from human Chk1 | | Descriptor: | ACETATE ION, GLYCEROL, cDNA FLJ56409, ... | | Authors: | Emptage, R.P, Marmorstein, R. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Intramolecular autoinhibition of checkpoint kinase 1 is mediated by conserved basic motifs of the C-terminal kinase-associated 1 domain.
J. Biol. Chem., 292, 2017
|
|
6C95
 
 | |
1IHB
 
 | | CRYSTAL STRUCTURE OF P18-INK4C(INK6) | | Descriptor: | CYCLIN-DEPENDENT KINASE 6 INHIBITOR | | Authors: | Ravichandran, V, Swaminathan, K, Marmorstein, R. | | Deposit date: | 1997-10-25 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the CDK4/6 inhibitory protein p18INK4c provides insights into ankyrin-like repeat structure/function and tumor-derived p16INK4 mutations.
Nat.Struct.Biol., 5, 1998
|
|
8F23
 
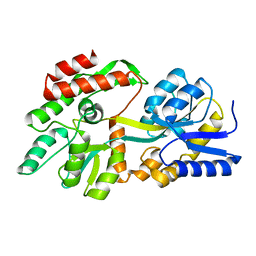 | | The crystal structure of a rationally designed zinc sensor based on maltose binding protein - Apo conformation | | Descriptor: | Zinc Sensor protein | | Authors: | Zhao, Z, Zhou, M, Zemerov, S.d, Marmorstein, R, Dmochowski, I.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rational design of a genetically encoded NMR zinc sensor.
Chem Sci, 14, 2023
|
|
8ETB
 
 | | the crystal structure of a rationally designed zinc sensor based on maltose binding protein - Zn binding conformation | | Descriptor: | ACETATE ION, ZINC ION, Zinc Sensor protein | | Authors: | Zhao, Z, Zhou, M, Zemerov, S.D, Marmorstein, R, Dmochowski, I.J. | | Deposit date: | 2022-10-16 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Rational design of a genetically encoded NMR zinc sensor.
Chem Sci, 14, 2023
|
|
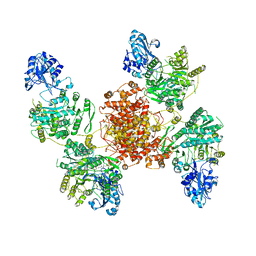
6UIA
 
 | |
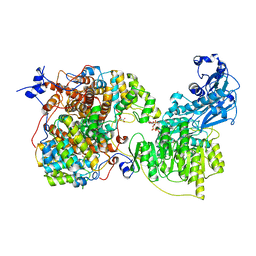
7LJ9
 
 | |
3BY4
 
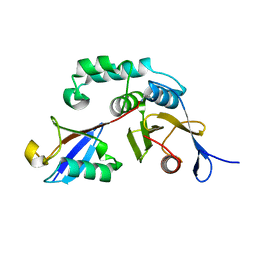 | |
3C0R
 
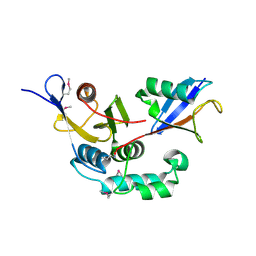 | |
1CM0
 
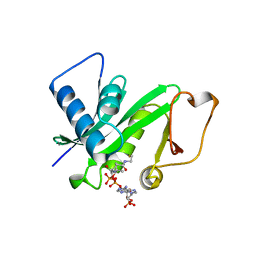 | | CRYSTAL STRUCTURE OF THE PCAF/COENZYME-A COMPLEX | | Descriptor: | COENZYME A, P300/CBP ASSOCIATING FACTOR | | Authors: | Clements, A, Rojas, J.R, Trievel, R.C, Wang, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-12 | | Release date: | 1999-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A.
EMBO J., 18, 1999
|
|
1HWT
 
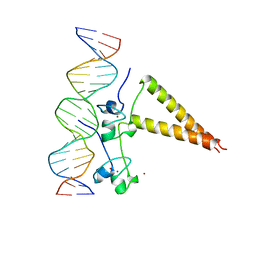 | | STRUCTURE OF A HAP1/DNA COMPLEX REVEALS DRAMATICALLY ASYMMETRIC DNA BINDING BY A HOMODIMERIC PROTEIN | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*C)-3'), PROTEIN (HEME ACTIVATOR PROTEIN), ... | | Authors: | King, D.A, Zhang, L, Guarente, L, Marmorstein, R. | | Deposit date: | 1998-09-17 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a HAP1-DNA complex reveals dramatically asymmetric DNA binding by a homodimeric protein.
Nat.Struct.Biol., 6, 1999
|
|
6C9M
 
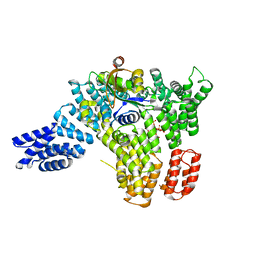 | |
5ICV
 
 | | Crystal structure of human NatF (hNaa60) bound to a bisubstrate analogue | | Descriptor: | MET-LYS-ALA-VAL-LIG, N-alpha-acetyltransferase 60, [5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)furan-2-yl]methyl (3R)-4-{[3-({(E)-2-[(2,2-dihydroxyethyl)sulfanyl]ethenyl}amino)-3-oxopropyl]amino}-3-hydroxy-2,2-dimethyl-4-oxobutyl dihydrogen diphosphate | | Authors: | Stove, S.I, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of the Golgi-Associated Human N alpha-Acetyltransferase 60 Reveals the Molecular Determinants for Substrate-Specific Acetylation.
Structure, 24, 2016
|
|
4KVX
 
 | | Crystal structure of Naa10 (Ard1) bound to AcCoA | | Descriptor: | ACETYL COENZYME *A, N-terminal acetyltransferase A complex catalytic subunit ard1 | | Authors: | Liszczak, G.P, Marmorstein, R.Q. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for N-terminal acetylation by the heterodimeric NatA complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
