3QJO
 
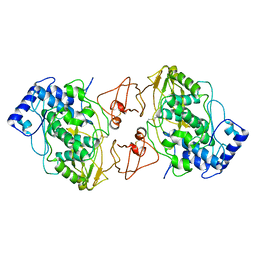 | | Refined Structure of the functional unit (KLH1-H) of keyhole limpet hemocyanin | | Descriptor: | COPPER (II) ION, Hemocyanin 1 | | Authors: | Jaenicke, E, Buchler, K, Decker, H, Markl, J, Schroder, G.F. | | Deposit date: | 2011-01-30 | | Release date: | 2011-05-25 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The refined structure of functional unit h of keyhole limpet hemocyanin (KLH1-h) reveals disulfide bridges.
Iubmb Life, 63, 2011
|
|
3L6W
 
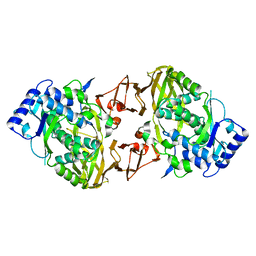 | | Structure of the collar functional unit (KLH1-H) of keyhole limpet hemocyanin | | Descriptor: | Hemocyanin 1 | | Authors: | Jaenicke, E, Buechler, K, Markl, J, Decker, H, Barends, T.R.M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Cupredoxin-like Domains in Hemocyanins.
Biochem.J., 2009
|
|
4BED
 
 | |
4AOE
 
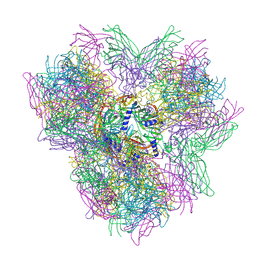 | | Biomphalaria glabrata Acetylcholine-binding protein type 2 (BgAChBP2) | | Descriptor: | ACETYLCHOLINE-BINDING PROTEIN TYPE 2 | | Authors: | Saur, M, Moeller, V, Kapetanopoulos, K, Braukmann, S, Gebauer, W, Tenzer, S, Markl, J. | | Deposit date: | 2012-03-26 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Acetylcholine-Binding Protein in the Hemolymph of the Planorbid Snail Biomphalaria Glabrata is a Pentagonal Dodecahedron (60 Subunits)
Plos One, 7, 2012
|
|
4AOD
 
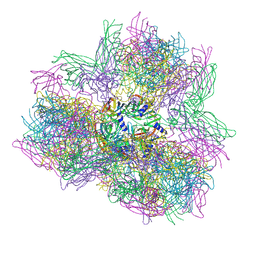 | | Biomphalaria glabrata Acetylcholine-binding protein type 1 (BgAChBP1) | | Descriptor: | ACETYLCHOLINE-BINDING PROTEIN TYPE 1 | | Authors: | Saur, M, Moeller, V, Kapetanopoulos, K, Braukmann, S, Gebauer, W, Tenzer, S, Markl, J. | | Deposit date: | 2012-03-26 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Acetylcholine-Binding Protein in the Hemolymph of the Planorbid Snail Biomphalaria Glabrata is a Pentagonal Dodecahedron (60 Subunits)
Plos One, 7, 2012
|
|
6NU4
 
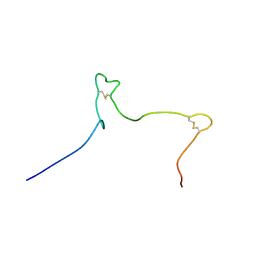 | | Solution structure of the Arabidopsis thaliana RALF8 peptide | | Descriptor: | Protein RALF-like 8 | | Authors: | Lee, W, Markley, J.L, Frederick, R.O, Miyoshi, H, Tonelli, M, Cornilescu, G, Cornilescu, C, Sussman, M.R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Function and solution structure of the Arabidopsis thaliana RALF8 peptide.
Protein Sci., 28, 2019
|
|
1YV8
 
 | |
6O6W
 
 | | Solution structure of human myeloid-derived growth factor | | Descriptor: | Myeloid-derived growth factor | | Authors: | Bortnov, V, Tonelli, M, Lee, W, Markley, J.L, Mosher, D.F. | | Deposit date: | 2019-03-07 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum.
Nat Commun, 10, 2019
|
|
1G11
 
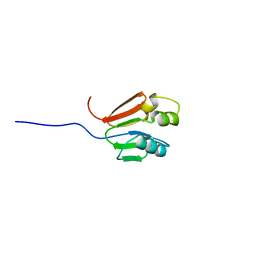 | | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR PROTEIN NMR STRUCTURE | | Descriptor: | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR | | Authors: | Hemmi, H, Studts, J.M, Chae, Y.K, Song, J, Markley, J.L, Fox, B.G. | | Deposit date: | 2000-10-10 | | Release date: | 2001-05-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the toluene 4-monooxygenase effector protein (T4moD).
Biochemistry, 40, 2001
|
|
1G10
 
 | | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR PROTEIN NMR STRUCTURE | | Descriptor: | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR | | Authors: | Hemmi, H, Studts, J.M, Chae, Y.K, Song, J, Markley, J.L, Fox, B.G. | | Deposit date: | 2000-10-10 | | Release date: | 2001-05-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the toluene 4-monooxygenase effector protein (T4moD).
Biochemistry, 40, 2001
|
|
5VSO
 
 | | NMR structure of Ydj1 J-domain, a cytosolic Hsp40 from Saccharomyces cerevisiae | | Descriptor: | Yeast dnaJ protein 1 | | Authors: | Ciesielski, S.J, Tonelli, M, Lee, W, Cornilescu, G, Markley, J.L, Schilke, B.A, Ziegelhoffer, T, Craig, E.A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Broadening the functionality of a J-protein/Hsp70 molecular chaperone system.
PLoS Genet., 13, 2017
|
|
6CGH
 
 | | Solution structure of the four-helix bundle region of human J-protein Zuotin, a component of ribosome-associated complex (RAC) | | Descriptor: | DnaJ homolog subfamily C member 2 | | Authors: | Shrestha, O.K, Lee, W, Tonelli, M, Cornilescu, G, Markley, J.L, Ciesielski, S.J, Craig, E.A. | | Deposit date: | 2018-02-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and evolution of the 4-helix bundle domain of Zuotin, a J-domain protein co-chaperone of Hsp70.
Plos One, 14, 2019
|
|
2QL0
 
 | | Zinc-substituted Rubredoxin from Desulfovibrio Vulgaris | | Descriptor: | Rubredoxin, ZINC ION | | Authors: | Goodfellow, B.J, Nunes, S.G, Volkman, B.F, Moura, J.G, Macedo, A.L, Duarte, I.C, Markley, J.L, Moura, I. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: |
To be published
|
|
1FRD
 
 | | MOLECULAR STRUCTURE OF THE OXIDIZED, RECOMBINANT, HETEROCYST (2FE-2S) FERREDOXIN FROM ANABAENA 7120 DETERMINED TO 1.7 ANGSTROMS RESOLUTION | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, HETEROCYST [2FE-2S] FERREDOXIN | | Authors: | Jacobson, B.L, Chae, Y.K, Markley, J.L, Rayment, I, Holden, H.M. | | Deposit date: | 1993-04-14 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular structure of the oxidized, recombinant, heterocyst [2Fe-2S] ferredoxin from Anabaena 7120 determined to 1.7-A resolution.
Biochemistry, 32, 1993
|
|
1FXA
 
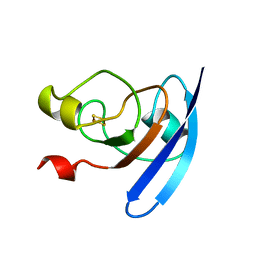 | | CRYSTALLIZATION AND STRUCTURE DETERMINATION TO 2.5-ANGSTROMS RESOLUTION OF THE OXIDIZED [2FE-2S] FERREDOXIN ISOLATED FROM ANABAENA 7120 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, [2FE-2S] FERREDOXIN | | Authors: | Rypniewski, W.R, Breiter, D.R, Benning, M.M, Wesenberg, G, Oh, B.-H, Markley, J.L, Rayment, I, Holden, H.M. | | Deposit date: | 1991-01-09 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and structure determination to 2.5-A resolution of the oxidized [2Fe-2S] ferredoxin isolated from Anabaena 7120.
Biochemistry, 30, 1991
|
|
7KNV
 
 | | Solution NMR structure of CDHR3 extracellular domain EC1 | | Descriptor: | CALCIUM ION, Cadherin-related family member 3 | | Authors: | Lee, W, Tonelli, M, Frederick, R.O, Watters, K.E, Markley, J.L, Palmenberg, A.C. | | Deposit date: | 2020-11-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Determination of the CDHR3 Rhinovirus-C Binding Domain, EC1
Viruses, 13, 2021
|
|
1M8B
 
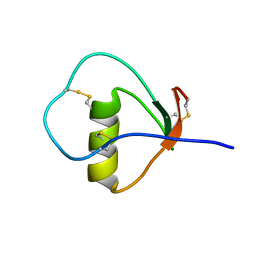 | | Solution structure of the C State of turkey ovomucoid at pH 2.5 | | Descriptor: | Ovomucoid | | Authors: | Song, J, Laskowski Jr, M, Qasim, M.A, Markley, J.L. | | Deposit date: | 2002-07-24 | | Release date: | 2002-09-04 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Two conformational states of Turkey ovomucoid third domain at low pH: three-dimensional structures, internal dynamics, and interconversion kinetics and thermodynamics.
Biochemistry, 42, 2003
|
|
1M8C
 
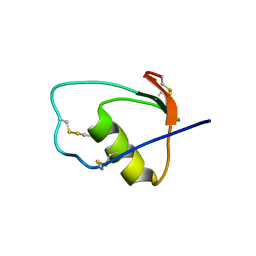 | | SOLUTION STRUCTURE OF THE T State OF TURKEY OVOMUCOID AT PH 2.5 | | Descriptor: | Ovomucoid | | Authors: | Song, J, Laskowski Jr, M, Qasim, M.A, Markley, J.L. | | Deposit date: | 2002-07-24 | | Release date: | 2002-09-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Two conformational states of Turkey ovomucoid third domain at low pH: three-dimensional structures, internal dynamics, and interconversion kinetics and thermodynamics.
Biochemistry, 42, 2003
|
|
1ZXF
 
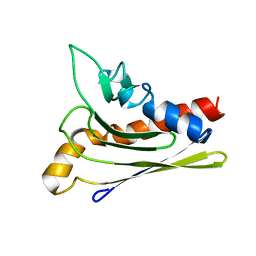 | | Solution structure of a self-sacrificing resistance protein, CalC from Micromonospora echinospora | | Descriptor: | CalC | | Authors: | Singh, S, Hager, M.H, Zhang, C, Griffith, B.R, Lee, M.S, Hallenga, K, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
1OMU
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (REFINED MODEL USING NETWORK EDITING ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
1OMT
 
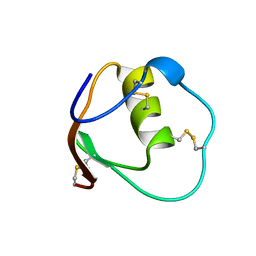 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (STANDARD NOESY ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
2BRZ
 
 | | SOLUTION NMR STRUCTURE OF THE SWEET PROTEIN BRAZZEIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BRAZZEIN | | Authors: | Caldwell, J.E, Abildgaard, F, Dzakula, Z, Ming, D, Hellekant, G, Markley, J.L. | | Deposit date: | 1998-04-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the thermostable sweet-tasting protein brazzein.
Nat.Struct.Biol., 5, 1998
|
|
2BAI
 
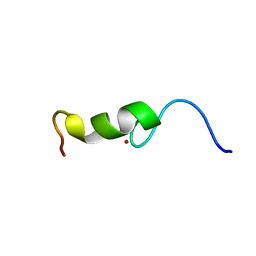 | | The Zinc finger domain of Mengovirus Leader polypeptide | | Descriptor: | Genome polyprotein, ZINC ION | | Authors: | Cornilescu, C.C, Porter, F.W, Qin, Z, Lee, M.S, Palmenberg, A.C, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the mengovirus Leader protein zinc-finger domain.
Febs Lett., 582, 2008
|
|
2LJL
 
 | |
2ARF
 
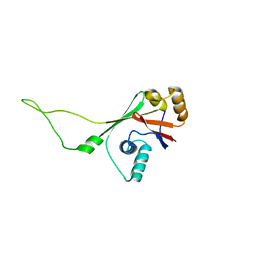 | | Solution structure of the Wilson ATPase N-domain in the presence of ATP | | Descriptor: | WILSON DISEASE ATPASE | | Authors: | Dmitriev, O, Tsivkovskii, R, Abildgaard, F, Morgan, C.T, Markley, J.L, Lutsenko, S. | | Deposit date: | 2005-08-19 | | Release date: | 2006-02-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-domain of Wilson disease protein: Distinct nucleotide-binding environment and effects of disease mutations
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
