2N3O
 
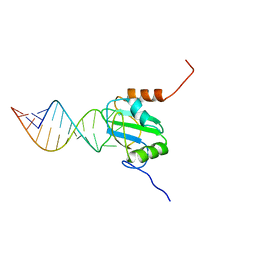 | | Structure of PTB RRM1(41-163) bound to an RNA stemloop containing a structured loop derived from viral internal ribosomal entry site RNA | | Descriptor: | Polypyrimidine tract-binding protein 1, RNA (5'-R(*GP*GP*GP*AP*CP*CP*UP*GP*GP*UP*CP*UP*UP*UP*CP*CP*AP*GP*GP*UP*CP*CP*C)-3') | | Authors: | Maris, C, Jayne, S.F, Damberger, F.F, Ravindranathan, S, Allain, F.H.-T. | | Deposit date: | 2015-06-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | C-terminal helix folding upon pyrimidine-rich hairpin binding to PTB RRM1. Implications for PTB function in Encephalomyocarditis virus IRES activity.
To be Published
|
|
1YNE
 
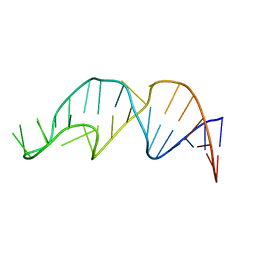 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | APOLIPOPROTEIN B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1YNG
 
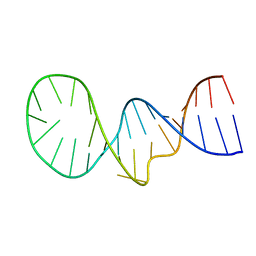 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1YLG
 
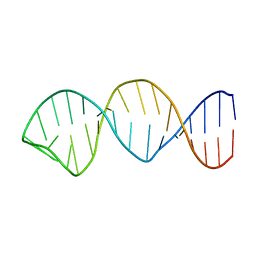 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1YNC
 
 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
2GIP
 
 | |
2GIO
 
 | |
5N8L
 
 | |
5N8M
 
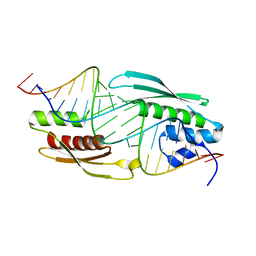 | |
2MZ1
 
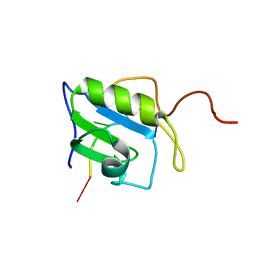 | | Solution structure of hnRNP C RRM in complex with 5'-UUUUC-3' RNA | | Descriptor: | 5'-R(*UP*UP*UP*UP*C)-3', Heterogeneous nuclear ribonucleoproteins C1/C2 | | Authors: | Cienikova, Z, Damberger, F.F, Hall, J, Allain, F.H.-T, Maris, C. | | Deposit date: | 2015-02-05 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into poly(uridine) tract recognition by the hnRNP C RNA recognition motif.
J.Am.Chem.Soc., 136, 2014
|
|
2MXY
 
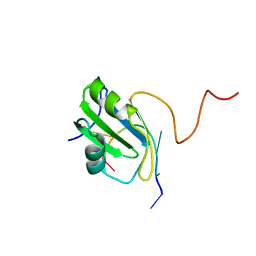 | | Solution structure of hnRNP C RRM in complex with 5'-AUUUUUC-3' RNA | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*C)-3', Heterogeneous nuclear ribonucleoproteins C1/C2 | | Authors: | Cienikova, Z, Damberger, F.F, Hall, J, Allain, F.H.-T, Maris, C. | | Deposit date: | 2015-01-19 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into poly(uridine) tract recognition by the hnRNP C RNA recognition motif.
J.Am.Chem.Soc., 136, 2014
|
|
9CR4
 
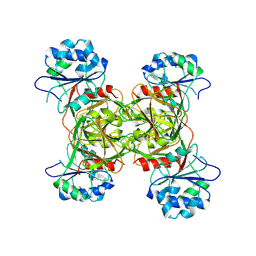 | | CryoEM Structure of the C-terminally truncated form of human NAD Kinase | | Descriptor: | NAD kinase | | Authors: | Li, Y, Chen, Z, Mary, C, Labesse, G, Hoxhaj, G. | | Deposit date: | 2024-07-20 | | Release date: | 2025-01-08 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure and regulation of human NAD kinase.
Sci Adv, 11, 2025
|
|
9CRA
 
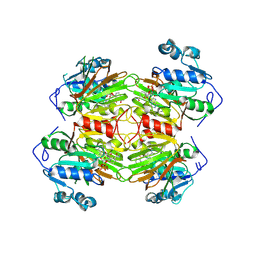 | | CryoEM Structure of the C-terminally truncated form of human NAD Kinase bound to NAD | | Descriptor: | NAD kinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, Y, Chen, Z, Mary, C, Labesse, G, Hoxhaj, G. | | Deposit date: | 2024-07-21 | | Release date: | 2025-01-08 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Cryo-EM structure and regulation of human NAD kinase.
Sci Adv, 11, 2025
|
|
4UYK
 
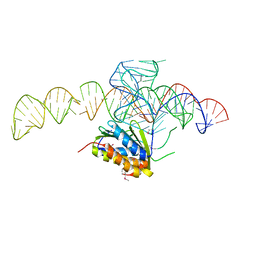 | | Crystal structure of a Signal Recognition Particle Alu domain in the elongation arrest conformation | | Descriptor: | SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, SRP RNA | | Authors: | Bousset, L, Mary, C, Brooks, M.A, Scherrer, A, Strub, K, Cusack, S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Crystal Structure of a Signal Recognition Particle Alu Domain in the Elongation Arrest Conformation.
RNA, 20, 2014
|
|
4UYJ
 
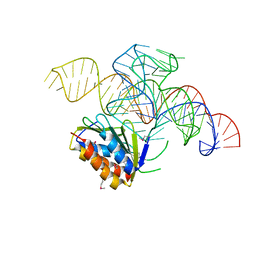 | | Crystal structure of a Signal Recognition Particle Alu domain in the elongation arrest conformation | | Descriptor: | SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, SRP RNA | | Authors: | Bousset, L, Mary, C, Brooks, M.A, Scherrer, A, Strub, K, Cusack, S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of a Signal Recognition Particle Alu Domain in the Elongation Arrest Conformation.
RNA, 20, 2014
|
|
7R4K
 
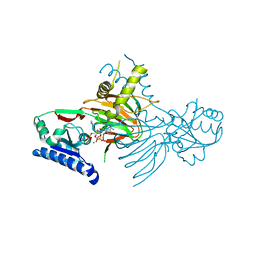 | | Crystal structure of human mitochondrial NAD kinase | | Descriptor: | MAGNESIUM ION, NAD kinase 2, mitochondrial, ... | | Authors: | Labesse, G, Mary, C, Gelin, M, Lionne, C. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Crystal structure of human NADK2 reveals a dimeric organization and active site occlusion by lysine acetylation.
Mol.Cell, 82, 2022
|
|
7R4J
 
 | | Crystal structure of human mitochondrial NAD kinase | | Descriptor: | CALCIUM ION, NAD kinase 2, mitochondrial | | Authors: | Labesse, G, Mary, C, Gelin, M, Lionne, C. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of human NADK2 reveals a dimeric organization and active site occlusion by lysine acetylation.
Mol.Cell, 82, 2022
|
|
7R4L
 
 | | Crystal structure of human mitochondrial NAD kinase | | Descriptor: | FE (III) ION, NAD kinase 2, mitochondrial, ... | | Authors: | Labesse, G, Mary, C, Gelin, M, Lionne, C. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human NADK2 reveals a dimeric organization and active site occlusion by lysine acetylation.
Mol.Cell, 82, 2022
|
|
7R4M
 
 | | Crystal structure of mitochondrial NAD kinase | | Descriptor: | CALCIUM ION, NAD kinase 2, mitochondrial, ... | | Authors: | Labesse, G, Mary, C, Gelin, M, Lionne, C. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of human NADK2 reveals a dimeric organization and active site occlusion by lysine acetylation.
Mol.Cell, 82, 2022
|
|
