5FUG
 
 | | Crystal structure of a human YL1-H2A.Z-H2B complex | | Descriptor: | HISTONE H2A.Z, HISTONE H2B TYPE 1-J, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 72 HOMOLOG | | Authors: | Latrick, C.M, Marek, M, Ouararhni, K, Papin, C, Stoll, I, Ignatyeva, M, Obri, A, Ennifar, E, Dimitrov, S, Romier, C, Hamiche, A. | | Deposit date: | 2016-01-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Basis and Specificity of H2A.Z-H2B Recognition and Deposition by the Histone Chaperone Yl1
Nat.Struct.Mol.Biol., 23, 2016
|
|
8AX8
 
 | |
8AQ6
 
 | |
8AQI
 
 | |
8AQH
 
 | | NanoLuc-Y94A luciferase mutant | | Descriptor: | NanoLuc luciferase | | Authors: | Nemergut, M, Marek, M. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Illuminating the mechanism and allosteric behavior of NanoLuc luciferase.
Nat Commun, 14, 2023
|
|
8AX9
 
 | |
8B5K
 
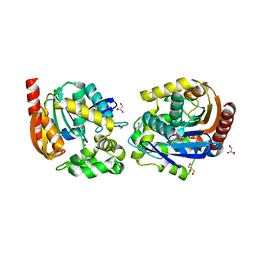 | | Structure of haloalkane dehalogenase DmmarA from Mycobacterium marinum at pH 6.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Snajdarova, K, Marek, M. | | Deposit date: | 2022-09-23 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Atypical homodimerization revealed by the structure of the (S)-enantioselective haloalkane dehalogenase DmmarA from Mycobacterium marinum.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
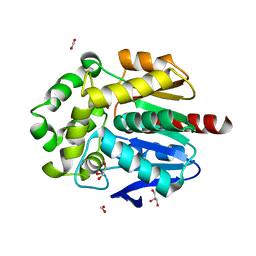
8B5O
 
 | |
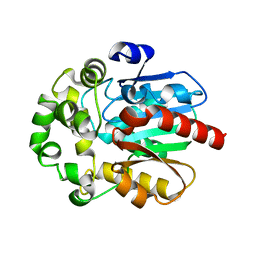
6S6E
 
 | |
7OMO
 
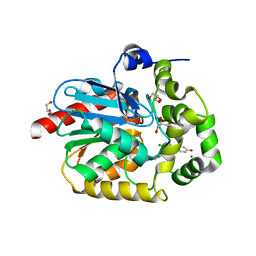 | |
7Q1C
 
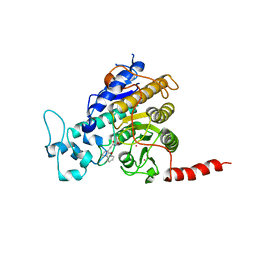 | | Crystal structure of Trypanosoma cruzi histone deacetylase DAC2 complexed with a hydroxamate inhibitor | | Descriptor: | (E)-3-dibenzofuran-4-yl-N-oxidanyl-prop-2-enamide, Histone deacetylase DAC2, POTASSIUM ION, ... | | Authors: | Ramos-Morales, E, Marek, M, Romier, C. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Species-selective targeting of pathogens revealed by the atypical structure and active site of Trypanosoma cruzi histone deacetylase DAC2.
Cell Rep, 37, 2021
|
|
6S97
 
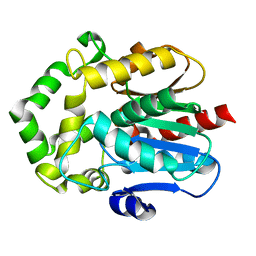 | |
6SP8
 
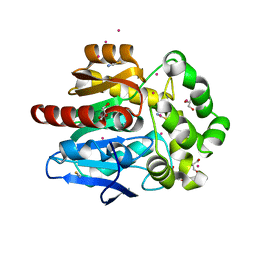 | | Structure of hyperstable haloalkane dehalogenase variant DhaA115 prepared by the 'soak-and-freeze' method under 150 bar of krypton pressure | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Chmelova, K, Markova, K, Damborsky, J, Marek, M. | | Deposit date: | 2019-08-31 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst.
Chem Sci, 11, 2020
|
|
6SP5
 
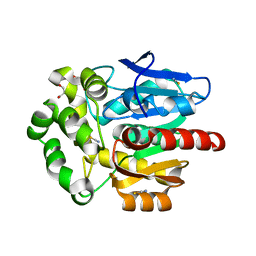 | | Structure of hyperstable haloalkane dehalogenase variant DhaA115 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Chmelova, K, Markova, K, Damborsky, J, Marek, M. | | Deposit date: | 2019-08-30 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst.
Chem Sci, 11, 2020
|
|
7OMR
 
 | |
7OMD
 
 | | Crystal structure of azacoelenterazine-bound Renilla reniformis luciferase variant RLuc8-D162A | | Descriptor: | 6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-8-(phenylmethyl)-[1,2,4]triazolo[4,3-a]pyrazin-3-one, CHLORIDE ION, Coelenterazine h 2-monooxygenase, ... | | Authors: | Schenkmayerova, A, Janin, Y.L, Marek, M. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Catalytic mechanism for Renilla-type luciferases
Nat Catal, 2023
|
|
6YN2
 
 | |
6XTC
 
 | |
6XT8
 
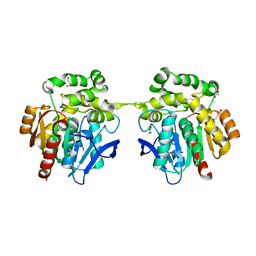 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Markova, K, Damborsky, J, Marek, M. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
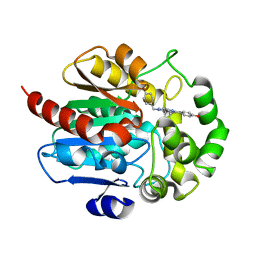
7OME
 
 | |
6Y9E
 
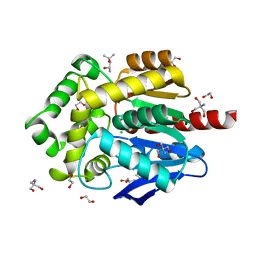 | | Crystal structure of putative ancestral haloalkane dehalogenase AncHLD2 (node 2) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Chaloupkova, R, Damborsky, J, Marek, M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of hyperstable ancestral haloalkane dehalogenases show restricted conformational dynamics.
Comput Struct Biotechnol J, 18, 2020
|
|
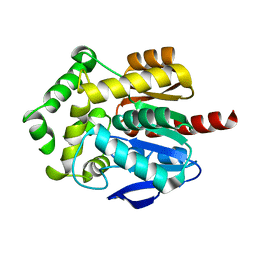
6Y9G
 
 | |
6Y9F
 
 | |
6GX3
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 1 | | Descriptor: | 4-chloranyl-~{N}-oxidanyl-1-benzothiophene-2-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
6GXA
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 2 | | Descriptor: | (~{E})-3-(2-chlorophenyl)-~{N}-oxidanyl-prop-2-enamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
