5I6A
 
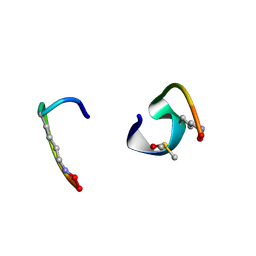 | | bicyclo[3.3.2]decapeptide | | Descriptor: | ALA-PHE-GLY-LYD-VAL-PHE-PRO-GLN-ALA-GLY, DIMETHYL SULFOXIDE | | Authors: | Bartoloni, M, Waltersperger, S, Bumann, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-09 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (0.813 Å) | | Cite: | Stereoselective synthesis and structure determination of a bicyclo[3.3.2]decapeptide
Arkivoc, 2014, 2014
|
|
3GVM
 
 | |
8BEB
 
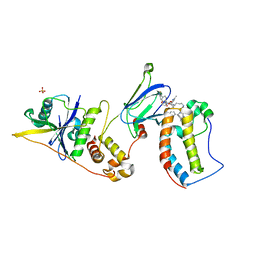 | | Ternary complex between VCB, BRD4-BD1 and PROTAC 49 | | Descriptor: | (2~{S},4~{R})-~{N}-[(1~{S})-3-[4-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]butylamino]-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-3-oxidanylidene-propyl]-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Elongin-B, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-21 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
8BDX
 
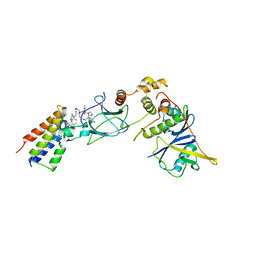 | | Ternary complex between VCB, BRD4-BD2 and PROTAC 48 | | Descriptor: | (2S,4R)-N-[(1S)-1-(4-chlorophenyl)-3-[2-[2-[2-[2-[2-[(9S)-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8$l^{5},11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethylamino]-3-oxidanylidene-propyl]-1-[(2R)-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Elongin-B, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-20 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
8BRF
 
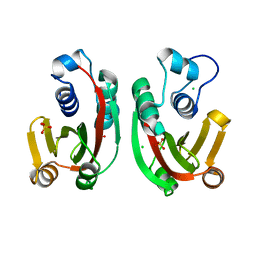 | |
8BS0
 
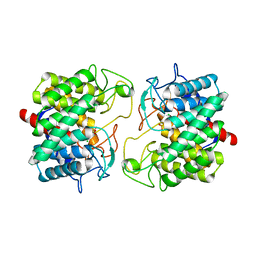 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at 80 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BRZ
 
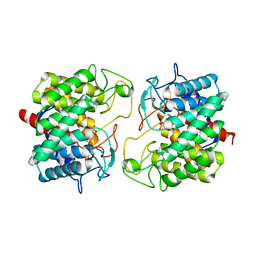 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at 52 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BRY
 
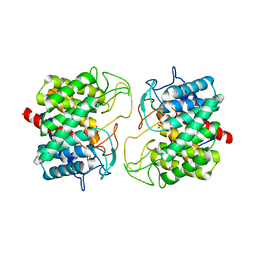 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at atmospheric pressure | | Descriptor: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
2R15
 
 | |
5TIQ
 
 | | The Structure of the Major Capsid protein of PBCV-1 | | Descriptor: | 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-beta-L-rhamnopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-D-mannopyranose-(1-3)-alpha-D-rhamnopyranose-(1-3)][alpha-D-galactopyranose-(1-2)]alpha-L-fucopyranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-glucopyranose, MERCURY (II) ION, Major capsid protein, ... | | Authors: | Klose, T, De Castro, C, Speciale, I, Molinaro, A, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structure of the chlorovirus PBCV-1 major capsid glycoprotein determined by combining crystallographic and carbohydrate molecular modeling approaches.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5K8J
 
 | | Structure of Caulobacter crescentus VapBC1 (apo form) | | Descriptor: | GLYCEROL, Ribonuclease VapC, VapB family protein | | Authors: | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | Deposit date: | 2016-05-30 | | Release date: | 2016-12-28 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
5HX2
 
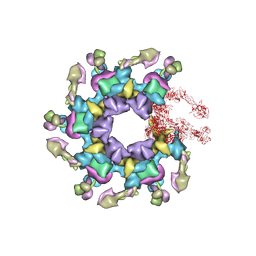 | | In vitro assembled star-shaped hubless T4 baseplate | | Descriptor: | Baseplate wedge protein gp10, Baseplate wedge protein gp53, Baseplate wedge protein gp6, ... | | Authors: | Yap, M.L, Klose, T, Fokine, A, Rossmann, M.G. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Role of bacteriophage T4 baseplate in regulating assembly and infection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8ATD
 
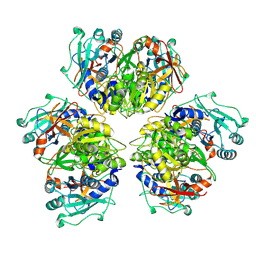 | | Wild type hexamer oxalyl-CoA synthetase (OCS) | | Descriptor: | Oxalate--CoA ligase | | Authors: | Lill, P, Burgi, J, Raunser, S, Wilmanns, M, Gatsogiannis, C. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol.Chem., 404, 2023
|
|
8AFG
 
 | |
8ATI
 
 | |
3ZNJ
 
 | | Crystal structure of unliganded ClcF from R.opacus 1CP in crystal form 1. | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
8BC6
 
 | |
3ZR4
 
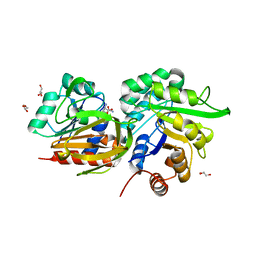 | | STRUCTURAL EVIDENCE FOR AMMONIA TUNNELING ACROSS THE (BETA-ALPHA)8 BARREL OF THE IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE BIENZYME COMPLEX | | Descriptor: | GLUTAMINE, GLYCEROL, IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISF, ... | | Authors: | Vega, M.C, Kuper, J, Haeger, M.C, Mohrlueder, J, Marquardt, S, Sterner, R, Wilmanns, M. | | Deposit date: | 2011-06-13 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Catalysis Uncoupling in a Glutamine Amidotransferase Bienzyme by Unblocking the Glutaminase Active Site.
Chem.Biol., 19, 2012
|
|
8RDS
 
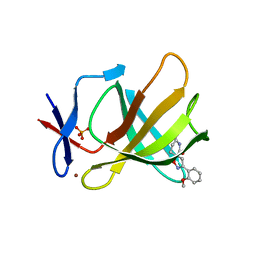 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8i | | Descriptor: | (5~{S})-3-(2-methoxyphenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDT
 
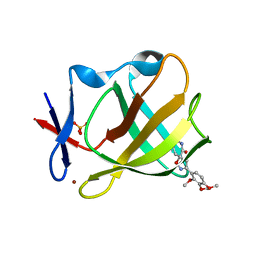 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8j | | Descriptor: | (5~{S})-3-(2,3,4-trimethoxyphenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDQ
 
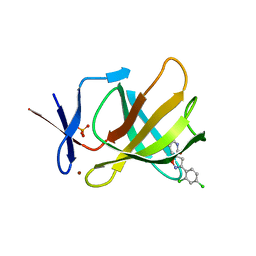 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8b | | Descriptor: | (5S)-3-(2,4-dichlorophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
6T7E
 
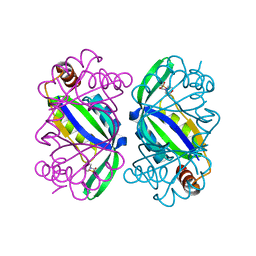 | | PII-like protein CutA from Nostoc sp. PCC7120 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Periplasmic divalent cation tolerance protein | | Authors: | Selim, K.A, Albrecht, R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 288, 2021
|
|
8AFV
 
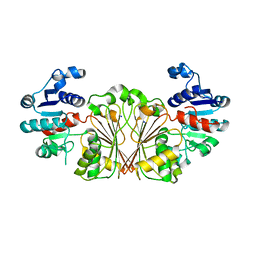 | | DaArgC3 - Engineered Formyl Phosphate Reductase with 3 substitutions (S178V, G182V, L233I) | | Descriptor: | N-acetyl-gamma-glutamyl-phosphate reductase, SODIUM ION | | Authors: | Pfister, P, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2022-07-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | N-acetyl-gamma-glutamyl-phosphate reductase of Denitrovibrio acetiphilus
To Be Published
|
|
8AFU
 
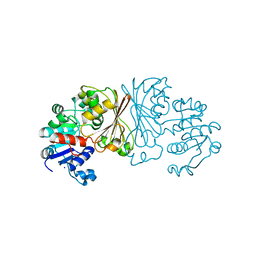 | |
5IRE
 
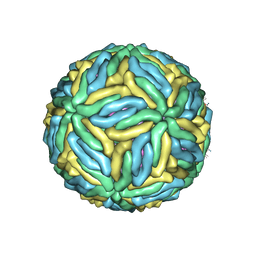 | | The cryo-EM structure of Zika Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein | | Authors: | Sirohi, D, Chen, Z, Sun, L, Klose, T, Pierson, T, Rossmann, M, Kuhn, R. | | Deposit date: | 2016-03-13 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom resolution cryo-EM structure of Zika virus.
Science, 352, 2016
|
|
